Extensive genome-wide autozygosity in the population isolates of Daghestan - PubMed (original) (raw)
Extensive genome-wide autozygosity in the population isolates of Daghestan
Tatiana M Karafet et al. Eur J Hum Genet. 2015 Oct.
Abstract
Isolated populations are valuable resources for mapping disease genes, as inbreeding increases genome-wide homozygosity and enhances the ability to map disease alleles on a genetically uniform background within a relatively homogenous environment. The populations of Daghestan are thought to have resided in the Caucasus Mountains for hundreds of generations and are characterized by a high prevalence of certain complex diseases. To explore the extent to which their unique population history led to increased levels of inbreeding, we genotyped >550 000 autosomal single-nucleotide polymorphisms (SNPs) in a set of 14 population isolates speaking Nakh-Daghestanian (ND) languages. The ND-speaking populations showed greatly elevated coefficients of inbreeding, very high numbers and long lengths of Runs of Homozygosity, and elevated linkage disequilibrium compared with surrounding groups from the Caucasus, the Near East, Europe, Central and South Asia. These results are consistent with the hypothesis that most ND-speaking groups descend from a common ancestral population that fragmented into a series of genetic isolates in the Daghestanian highlands. They have subsequently maintained a long-term small effective population size as a result of constant inbreeding and very low levels of gene flow. Given these findings, Daghestanian population isolates are likely to be useful for mapping genes associated with complex diseases.
Figures
Figure 1
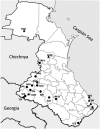
Approximate geographic location of sampling sites. Of the 20 Daghestanian populations sampled here, 14 speak unique languages that are part of the ND language family. Of the remaining six populations (non-ND), three speak languages that are closely related to the Turkic branch of Altaic language family (Kumyks, Nogais and Azerbaijans), and three speak languages belonging to the Iranian language branch of the Indo-European language family (ethnic Tats, Mountain Jews and a group of Azerbaijans originated in Iran). See Supplementary Table 1 for population codes. ND and non-ND populations are shown in circle and triangle, respectively.
Figure 2
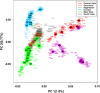
PCA analysis using the ‘drop one in' technique for 56 populations. Circled positions indicate the median coordinate values for populations. See Supplementary Table 1 for population codes.
Figure 3
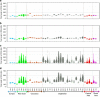
Distribution of total length of different ROH classes per individual in 40 populations. A box plot and a kernel density plot are shown as ‘violin plots' for classes A (a), B (b), C (c) and A, B, C classes pooled together (d). The white horizontal line is the median, whereas the black bar represents the interquartile range. Populations are grouped according to geographic regions.
Figure 4
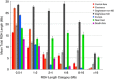
Regional distribution of ROH. The mean total ROH length (Mb)±SE is plotted for each geographic region.
Figure 5
The harmonic mean of _N_e for each population with sample size _N_≥10 over 50 recombination distance classes between 0.005 and 0.25 cM with increment 0.005 cM. The SEs for _N_e indicated by the bars were derived from separate analysis of each autosomal chromosome. Yorubans were included for comparisons with published data.
Similar articles
- Genomic patterns of homozygosity in worldwide human populations.
Pemberton TJ, Absher D, Feldman MW, Myers RM, Rosenberg NA, Li JZ. Pemberton TJ, et al. Am J Hum Genet. 2012 Aug 10;91(2):275-92. doi: 10.1016/j.ajhg.2012.06.014. Am J Hum Genet. 2012. PMID: 22883143 Free PMC article. - Age-based partitioning of individual genomic inbreeding levels in Belgian Blue cattle.
Solé M, Gori AS, Faux P, Bertrand A, Farnir F, Gautier M, Druet T. Solé M, et al. Genet Sel Evol. 2017 Dec 22;49(1):92. doi: 10.1186/s12711-017-0370-x. Genet Sel Evol. 2017. PMID: 29273000 Free PMC article. - Runs of homozygosity and inbreeding in thyroid cancer.
Thomsen H, Chen B, Figlioli G, Elisei R, Romei C, Cipollini M, Cristaudo A, Bambi F, Hoffmann P, Herms S, Landi S, Hemminki K, Gemignani F, Försti A. Thomsen H, et al. BMC Cancer. 2016 Mar 16;16:227. doi: 10.1186/s12885-016-2264-7. BMC Cancer. 2016. PMID: 26984635 Free PMC article. - Runs of homozygosity: current knowledge and applications in livestock.
Peripolli E, Munari DP, Silva MVGB, Lima ALF, Irgang R, Baldi F. Peripolli E, et al. Anim Genet. 2017 Jun;48(3):255-271. doi: 10.1111/age.12526. Epub 2016 Dec 1. Anim Genet. 2017. PMID: 27910110 Review. - Discovery of rare homozygous mutations from studies of consanguineous pedigrees.
Alkuraya FS. Alkuraya FS. Curr Protoc Hum Genet. 2012 Oct;Chapter 6:Unit6.12. doi: 10.1002/0471142905.hg0612s75. Curr Protoc Hum Genet. 2012. PMID: 23074070 Review.
Cited by
- Characterization of Danube Swabian population samples on a high-resolution genome-wide basis.
Bánfai Z, Kövesdi E, Sümegi K, Büki G, Szabó A, Magyari L, Ádám V, Pálos F, Miseta A, Kásler M, Melegh B. Bánfai Z, et al. BMC Genomics. 2023 Jan 9;24(1):9. doi: 10.1186/s12864-022-09092-5. BMC Genomics. 2023. PMID: 36624381 Free PMC article. - Inter-individual genomic heterogeneity within European population isolates.
Anagnostou P, Dominici V, Battaggia C, Lisi A, Sarno S, Boattini A, Calò C, Francalacci P, Vona G, Tofanelli S, Vilar MG, Colonna V, Pagani L, Destro Bisol G. Anagnostou P, et al. PLoS One. 2019 Oct 9;14(10):e0214564. doi: 10.1371/journal.pone.0214564. eCollection 2019. PLoS One. 2019. PMID: 31596857 Free PMC article. - The Effect of Consanguinity on Between-Individual Identity-by-Descent Sharing.
Severson AL, Carmi S, Rosenberg NA. Severson AL, et al. Genetics. 2019 May;212(1):305-316. doi: 10.1534/genetics.119.302136. Epub 2019 Mar 29. Genetics. 2019. PMID: 30926583 Free PMC article. - Inbreeding estimates in human populations: Applying new approaches to an admixed Brazilian isolate.
Lemes RB, Nunes K, Carnavalli JEP, Kimura L, Mingroni-Netto RC, Meyer D, Otto PA. Lemes RB, et al. PLoS One. 2018 Apr 24;13(4):e0196360. doi: 10.1371/journal.pone.0196360. eCollection 2018. PLoS One. 2018. PMID: 29689090 Free PMC article. - Relationship between Deleterious Variation, Genomic Autozygosity, and Disease Risk: Insights from The 1000 Genomes Project.
Pemberton TJ, Szpiech ZA. Pemberton TJ, et al. Am J Hum Genet. 2018 Apr 5;102(4):658-675. doi: 10.1016/j.ajhg.2018.02.013. Epub 2018 Mar 15. Am J Hum Genet. 2018. PMID: 29551419 Free PMC article.
References
- 1Acevedo-Whitehouse K, Gulland F, Greig D, Amos W: Inbreeding: disease susceptibility in California sea lions. Nature 2003; 422: 35. - PubMed
- 2Bittles A: Consanguinity and its relevance to clinical genetics. Clin Genet 2001; 60: 89–98. - PubMed
- 5Charlesworth D, Willis JH: The genetics of inbreeding depression. Nat Rev Genet 2009; 10: 783–796. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources