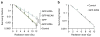Numerical chromosomal instability mediates susceptibility to radiation treatment - PubMed (original) (raw)
Lilian Kabeche 2, Matthew D Wood 3, Christopher D Laucius 2, Dian Qu 4, Ashley M Laughney 5, Gloria E Reynolds 6, Raymond J Louie 6, Joanna Phillips 4, Denise A Chan 6, Bassem I Zaki 7, John P Murnane 6, Claudia Petritsch 4, Duane A Compton 2
Affiliations
- PMID: 25606712
- PMCID: PMC4516720
- DOI: 10.1038/ncomms6990
Numerical chromosomal instability mediates susceptibility to radiation treatment
Samuel F Bakhoum et al. Nat Commun. 2015.
Abstract
The exquisite sensitivity of mitotic cancer cells to ionizing radiation (IR) underlies an important rationale for the widely used fractionated radiation therapy. However, the mechanism for this cell cycle-dependent vulnerability is unknown. Here we show that treatment with IR leads to mitotic chromosome segregation errors in vivo and long-lasting aneuploidy in tumour-derived cell lines. These mitotic errors generate an abundance of micronuclei that predispose chromosomes to subsequent catastrophic pulverization thereby independently amplifying radiation-induced genome damage. Experimentally suppressing whole-chromosome missegregation reduces downstream chromosomal defects and significantly increases the viability of irradiated mitotic cells. Further, orthotopically transplanted human glioblastoma tumours in which chromosome missegregation rates have been reduced are rendered markedly more resistant to IR, exhibiting diminished markers of cell death in response to treatment. This work identifies a novel mitotic pathway for radiation-induced genome damage, which occurs outside of the primary nucleus and augments chromosomal breaks. This relationship between radiation treatment and whole-chromosome missegregation can be exploited to modulate therapeutic response in a clinically relevant manner.
Conflict of interest statement
Competing financial interests: The authors declare no competing financial interests.
Figures
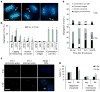
Figure 1. Ionizing radiation (IR) leads to numerical chromosomal instability
(a) Examples of U251 cells fixed 25 min after exposure to 12 Gy and exhibiting lagging chromosomes (LC), chromatin bridges (CB), acentric chromatin (AC) or a combination (LC +AC). Cells were stained for centromeres (green) and DNA (blue). Scale bar, 5 μm (b) Percentage of chromosome missegregation in anaphase spindles of RPE1, HCT116 and U251 cells as a function of IR dose. Bars represents mean±s.e.m., n = 150 cells, three experiments, *P<0.01, two-tailed _t_-test. (c) Examples of HCT116 nuclei stained for DNA (blue), centromere (red) and telomere (green) probes for human chromosome 2. White arrow denotes an aneuploid nucleus containing three copies of chromosome 2. Scale bar, 10 μm. (d) Per cent HCT116 nuclei containing whole-chromosome and segmental aneuploidy for chromosome 2. n = 300 cells, *P<0.05. (e) Percentage of chromosome missegregation in anaphase spindles of HCT116 p53 −/− cells exposed to 0 Gy (top) or 6 Gy (bottom) as a function of time after irradiation (mo, months).
Figure 2. Ionizing radiation (IR) induces chromosome segregation errors in vivo
(a) Schema for experiments depicted in b, c. Gy, gray; H&E, haematoxylin and eosin; SC, sub-cutaneous. (b) Example of H&E-stained SC-HCT116 p53 −/− xenografts showing normal anaphase and anaphase cells containing lagging chromosomes. Scale bar, 5 μm, inset bar, 1 μm. (c) Percentage of anaphase cells exhibiting lagging chromosomes in SC-HCT116 p53 −/− xenografts as a function of radiation dose. Bars represent mean±s.d., n = 4 mice; ****P<0.0001, two-tailed _t_-test. (d) Schema for experiments depicted in e. (e) Karyotype distribution of cells derived from six SC-HCT116 p53 −/− xenografts, each histogram represents a 100 spreads derived from a single tumour, two-tailed _t_-test.
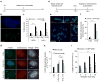
Figure 3. IR-induced chromosome segregation errors lead to widespread chromosomal damage
(a) Experimental schema for assessing the generation of IR-induced micronuclei. (b) Example image of a U251 cell containing a micronucleus stained for centromeres (green) and DNA (blue), scale bar, 5 μm. (c) Percentage of RPE1 and U251 cells containing micronuclei as a function of IR dose. Bars represents mean±s.e.m., n = 266–824 cells, three experiments, *P<0.05, **_P_<0.001, two-tailed _t_-test. (**d**) Experimental schema for assessing the generation of IR-induced chromosome pulverization. (**e**) Representative mitotic spread containing pulverized chromosomes from U251 cells irradiated with 12 Gy 24 h prior. Scale bar, 20 μm, which show a normal appearing chromosome (1), chromosome fragments (2–4), a dicentric chromosome (5) and uncondensed chromatin (6). (**f**) Percentage of mitotic spreads from containing pulverized chromosomes from control and U251 cells expressing GFP-Kif2b. Bars represent mean±s.e.m.; _n_>450 mitotic spreads, three experiments, *P<0.05, **_P_<0.001, two-tailed _t_-test. (**g**) Examples of U251 cells, exposed to 0 Gy, 6 Gy and fixed either 0.3 or 12 h later, containing micronuclei that encompass whole chromosomes (arrows) as evidenced by centromere staining (red) that were also stained for γ-H2AX (green) and DNA (blue). Scale bar, 5 μm (**h**) γ-H2AX fluorescence intensity in primary nuclei and micronuclei in U251 cells exposed to 0 and 12 Gy stained 0.3 or 12 h after irradiation. AU, arbitrary units; bars represent mean±s.e.m.; _n_>30 cells, three experiments, *P<0.05, **P<0.001, two-tailed _t_-test. (i) Percentage of anaphase spindles containing lagging chromosomes as a function of IR dose, in control and GFP-Kif2b-overexpressing U251 cells. Bars represents mean±s.e.m., n = 150 cells, three experiments, **P<0.001, two-tailed _t_-test.
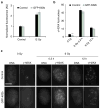
Figure 4. Kif2b overexpression does not alter IR-induced DNA breaks or repair
(a) Normalized fluorescence intensity of γ-H2AX staining during mitosis in control and GFP-Kif2b-overexpressing U251 cells. Bars represent mean±s.e.m. (b) The average number of γ-H2AX foci per nucleus as a function of IR dose 20 min and 12 h after IR exposure of control and GFP-Kif2b-overexpressing U251 cells. Bars represent mean±s.e.m., n = 52–151 cells; **P<0.005, two-tailed _t_-test. (c) Examples of cells irradiated with 0 or 6 Gy stained for DNA (left) and γ-H2AX right. Scale bar, 5 μm.
Figure 5. Chromosome segregation errors alter the viability of irradiated mitotic cells
Surviving fraction of irradiated mitotically enriched (a) and non-synchronized (b) U251 cells as well as cells overexpressing GFP, GFP-Kif2a, GFP-MCAK or GFP-Kif2b. Circles denote mean±s.e.m., n = 3 experiments, ***P<0.005, two-tailed _t_-test.
Figure 6. Reducing chromosome segregation errors induces radiation resistance in vivo
(a) Experimental schema for assessing in vivo tumour resistance; IC, intracranial. (b) Examples of bioluminescence images of mice harbouring IC GFP- and GFP-Kif2b-expressing tumours at day 0 (start of radiation treatment) and day 13 (end of radiation treatment). (c) Normalized bioluminescence signal overtime after initiation of IR treatment in IC GFP- and GFP-Kif2b-expressing U251 xenografts. Circles represent mean±s.e.m., n = 10 and 9 mice for GFP and GFP-Kif2b groups, respectively; *P<0.05, ***_P_<0.005, ****_P_<0.0001, two-tailed _t_-test. (**d**) Examples of haematoxylin and eosin-stained tumours expressing GFP or GFP-Kif2b, black arrows denote post-treatment tumours; scale bar, 500 μm. (**e**, **f**) Example of Ki67-stained specimens from irradiated tumours (**e**) and per cent Ki67-positive cells (**f**) in GFP- and GFP-Kif2b-expressing tumours. Bars represent mean±s.e.m., _n_ = 2–4 mice (UTx) and 7–9 mice (Tx), 1,080–2,511 cells per tumour; scale bar, 100 μm. (**g**) Mitotic count (per 10 high-power fields) in treated (Tx) and untreated (UTx) GFP- and GFP-Kif2b-expressing U251 xenografts. Bars represent mean±s.e.m., _n_ = 2–4 mice (UTx) and 7–9 mice (Tx). (**h**, **i**) Per cent of atypical mitotic cells (representative images depicted in h), in GFP- and GFP-Kif2b-expressing tumours. Bars represent mean±s.e.m., _n_ = 2–4 mice (UTx) and 7–9 mice (Tx). Scale bar, 7 μm. (**j**, **k**) Example of cleaved caspase 3 (CC3)-stained specimens from irradiated tumours (**j**), and semi-quantitative CC3 staining score in tumours (**k**), 1 + (<0.5% CC3-positive cells), 2 + (0.5–3%), 3 + (>3%); *P = 0.07, _χ2_-test, n = 2–4 mice (UTx) and 7–9 mice (Tx), 1,080–2,511 cells per tumour; scale bar, 500 μm. (l) Schematic diagram linking IR to chromosome segregation errors and downstream chromosomal structural defects.
Similar articles
- Folic acid deficiency increases chromosomal instability, chromosome 21 aneuploidy and sensitivity to radiation-induced micronuclei.
Beetstra S, Thomas P, Salisbury C, Turner J, Fenech M. Beetstra S, et al. Mutat Res. 2005 Oct 15;578(1-2):317-26. doi: 10.1016/j.mrfmmm.2005.05.012. Epub 2005 Jul 11. Mutat Res. 2005. PMID: 16005909 - DNA breaks and chromosome pulverization from errors in mitosis.
Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. Crasta K, et al. Nature. 2012 Jan 18;482(7383):53-8. doi: 10.1038/nature10802. Nature. 2012. PMID: 22258507 Free PMC article. - Mild replication stress causes aneuploidy by deregulating microtubule dynamics in mitosis.
Böhly N, Kistner M, Bastians H. Böhly N, et al. Cell Cycle. 2019 Oct;18(20):2770-2783. doi: 10.1080/15384101.2019.1658477. Epub 2019 Aug 25. Cell Cycle. 2019. PMID: 31448675 Free PMC article. - Deregulation of the centrosome cycle and the origin of chromosomal instability in cancer.
Lingle WL, Lukasiewicz K, Salisbury JL. Lingle WL, et al. Adv Exp Med Biol. 2005;570:393-421. doi: 10.1007/1-4020-3764-3_14. Adv Exp Med Biol. 2005. PMID: 18727509 Review. - Recent insights into the causes and consequences of chromosome mis-segregation.
Devillers R, Dos Santos A, Destombes Q, Laplante M, Elowe S. Devillers R, et al. Oncogene. 2024 Oct;43(43):3139-3150. doi: 10.1038/s41388-024-03163-5. Epub 2024 Sep 15. Oncogene. 2024. PMID: 39278989 Review.
Cited by
- Genomic profiling of newly diagnosed glioblastoma patients and its potential for clinical utility - a prospective, translational study.
Nørøxe DS, Yde CW, Østrup O, Michaelsen SR, Schmidt AY, Kinalis S, Torp MH, Skjøth-Rasmussen J, Brennum J, Hamerlik P, Poulsen HS, Nielsen FC, Lassen U. Nørøxe DS, et al. Mol Oncol. 2020 Nov;14(11):2727-2743. doi: 10.1002/1878-0261.12790. Epub 2020 Sep 18. Mol Oncol. 2020. PMID: 32885540 Free PMC article. - Radiosensitization-Related Cuproptosis LncRNA Signature in Non-Small Cell Lung Cancer.
Xu Q, Liu T, Wang J. Xu Q, et al. Genes (Basel). 2022 Nov 9;13(11):2080. doi: 10.3390/genes13112080. Genes (Basel). 2022. PMID: 36360316 Free PMC article. - DNA Damage Responses during the Cell Cycle: Insights from Model Organisms and Beyond.
Clay DE, Fox DT. Clay DE, et al. Genes (Basel). 2021 Nov 25;12(12):1882. doi: 10.3390/genes12121882. Genes (Basel). 2021. PMID: 34946831 Free PMC article. Review. - Radiotherapy is associated with a deletion signature that contributes to poor outcomes in patients with cancer.
Kocakavuk E, Anderson KJ, Varn FS, Johnson KC, Amin SB, Sulman EP, Lolkema MP, Barthel FP, Verhaak RGW. Kocakavuk E, et al. Nat Genet. 2021 Jul;53(7):1088-1096. doi: 10.1038/s41588-021-00874-3. Epub 2021 May 27. Nat Genet. 2021. PMID: 34045764 Free PMC article. - Mechanisms of chromosomal instability (CIN) tolerance in aggressive tumors: surviving the genomic chaos.
Dhital B, Rodriguez-Bravo V. Dhital B, et al. Chromosome Res. 2023 Apr 14;31(2):15. doi: 10.1007/s10577-023-09724-w. Chromosome Res. 2023. PMID: 37058263 Free PMC article. Review.
References
- Gunderson LL, Tepper JE. Clinical Radiation Oncology. Churchill Livingstone; 2011.
- Glass WA, Varma MN. Physical and Chemical Mechanisms in Molecular Radiation Biology. Springer; 2012.
- Chapman JR, Taylor MRG, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497–510. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 NS080619/NS/NINDS NIH HHS/United States
- P01 CA118816/CA/NCI NIH HHS/United States
- L60 MD001952/MD/NIMHD NIH HHS/United States
- R01NS080619/NS/NINDS NIH HHS/United States
- R37GM051542/GM/NIGMS NIH HHS/United States
- P30 CA023108/CA/NCI NIH HHS/United States
- R01 CA164746/CA/NCI NIH HHS/United States
- R01GM008704/GM/NIGMS NIH HHS/United States
- R37 GM051542/GM/NIGMS NIH HHS/United States
- T32 GM008704/GM/NIGMS NIH HHS/United States
- R01CA164746/CA/NCI NIH HHS/United States
- R01CA120205/CA/NCI NIH HHS/United States
- R01 CA120205/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical