Proteomic analyses reveal distinct chromatin-associated and soluble transcription factor complexes - PubMed (original) (raw)
Proteomic analyses reveal distinct chromatin-associated and soluble transcription factor complexes
Xu Li et al. Mol Syst Biol. 2015.
Abstract
The current knowledge on how transcription factors (TFs), the ultimate targets and executors of cellular signalling pathways, are regulated by protein-protein interactions remains limited. Here, we performed proteomics analyses of soluble and chromatin-associated complexes of 56 TFs, including the targets of many signalling pathways involved in development and cancer, and 37 members of the Forkhead box (FOX) TF family. Using tandem affinity purification followed by mass spectrometry (TAP/MS), we performed 214 purifications and identified 2,156 high-confident protein-protein interactions. We found that most TFs form very distinct protein complexes on and off chromatin. Using this data set, we categorized the transcription-related or unrelated regulators for general or specific TFs. Our study offers a valuable resource of protein-protein interaction networks for a large number of TFs and underscores the general principle that TFs form distinct location-specific protein complexes that are associated with the different regulation and diverse functions of these TFs.
Keywords: forkhead box; mass spectrometry; protein–protein interaction; transcriptional factor.
© 2015 The Authors. Published under the terms of the CC BY 4.0 license.
Figures
Figure 1
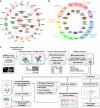
Proteomic analysis of human transcription factors
- Disease correlation of 19 TFs and 4 well-studied FOX family members, based on their GO annotations. Each colour indicates one disease. The size of each coloured pie indicates the relative ratio of –log (_P_-value) of GO annotations in the corresponding disease.
- Pathway correlation and structural superclasses of TFs. Each coloured area indicates one superfamily.
- Schematic diagram showing the major steps involved in TAP/MS screening and data analysis of human TFs and snapshot for each part of the data. Fifty-six transcription factors, together with 70 unrelated control proteins and control vector, were constructed into a vector harbouring a C-terminal SFB-tag through gateway technology. 293T cells stably expressing each bait protein were generated by stable transfection and puromycin selection. Protein was collected and separated into two fractions by a two-step lysis process. Through the standard tandem affinity purification steps, purified protein complexes were identified by mass spectrometry analysis, and final interacting proteins were generated by SAINT algorithm-based filtration. The data were subjected to prey functional categories analysis, interaction validation and function validation. Snapshots of data generated by each step were shown aside.
Figure 2
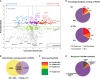
Proteomic analysis of human transcription factors and data validation
- Comparative analysis of prey specificities of TFs over different fractions. The _y_-axis shows TF-binding specificities of preys: positive, specifically associate with transcription factors group; negative, no binding preference. The _x_-axis depicts fraction specificities of preys: positive and negative numbers indicate preference for their enrichment in chromatin and soluble fractions, respectively. The size of a coloured bubble indicates the log (overall abundance) of individual preys. The selected preys were categorized into four groups based on their positions highlighted with different colours: red, specific co-regulators of TFs that may be involved in transcriptional regulation; purple, regulators with no fractional preference; blue, transcription-unrelated functions or negative regulators of TFs; green, potential regulators with less specificity; plus a group of abundant proteins with no binding preference, which were shown at the bottom of the map (grey).
- Data filtration using a modified SAINT algorithm. The total peptide and protein numbers obtained from mass spectrometry analysis are listed. The SAINT score > 0.80 was used as the cut-off to identify HCIPs, as suggested by the SAINT method. We also applied another filtration using the prey information in 70 control purifications to remove the non-specific bindings or contaminants. The numbers of HCIPs remaining after these two filtrations are shown here.
- Data reproducibility test based on biological replicates. The HCIP overlap ratio rises with the peptide numbers.
- Summary of HCIPs that overlap with those reported in knowledge PPI databases. 389 interactions of the total 2,156 HCIPs (˜18%) were reported previously.
- Summary of reciprocal purifications of 16 interactions identified from MAX, NFATC1, RBPJ and CREB1 purifications performed with the same TAP/MS protocol. 14 out of 16 preys captured their corresponding baits from reciprocal AP/MS, 13 of which are in the corresponding fractions.
Figure 3
Transcription factors form distinct complexes on and off chromatin
- HCIPs with highest spectra counts were listed. The length of each box with the protein name on it indicates the protein size. Black fonts indicate new interactions identified by our purifications. Orange fonts indicate interactions defined by our purifications and the literature.
- Total spectra counts of TFs in different fractions. The _y_-axis indicates the total spectra counts (TSC) of HCIPs in corresponding TF purifications. Red bar: TSC of HCIPs in chromatin fractions; blue bar: TSC of HCIPs in soluble fractions.
Figure 4
Protein categories and functions are different on and off chromatin
- Overlap Venn diagram of HCIPs in chromatin and soluble fractions. Only 196 HCIPs appear in both chromatin and soluble fractions, 120 of which are bait self-identifications.
- Function categories of HCIPs in the two fractions.
- Ubiquitin-related HCIPs enrichment in the two fractions. The _y_-axis indicates the number of HCIPs.
- FOXM1 HCIPs form distinct complexes in chromatin versus soluble fractions. HCIPs with highest spectra counts or previously known associated proteins were listed. Black font indicates the HCIPs defined by our purifications; grey font indicates the proteins defined by our purifications that form complexes with HCIPs, but are not in the HCIP list. Lines indicate the interactions defined in the literature. Prey dots in different colours indicate different function complexes defined in the literature.
- GO annotation of FOXM1 in molecular and cellular functions based on its HCIPs identified in chromatin or soluble fractions. Colours indicate the –log (_P_-value) of GO annotations.
- A model showing on/off chromatin functions of FOXM1 in mitosis and cell cycle progression. All of the components indicated were identified from FOXM1 purifications.
Figure 5
Functional validation of FOXN2 based on its interacting proteins in soluble and chromatin fractions
- FOXN2 HCIPs form distinct complexes in chromatin versus soluble fractions. The size of prey dots indicates the estimated abundance of preys. Lines indicate the interactions defined in the literature. CUL1 was identified in a parallel virus-based FOXN2 purification.
- 293T cells were transfected with constructs encoding MYC-tagged RFX1 and SFB-tagged FOXN2 or its DNA binding-defective mutant FOXN2 (H162R) as indicated. Pull-down experiments were carried out with S-protein beads and immunoblotted with antibodies as indicated.
- Overlap Venn diagram of FOXN2 and RFX1 target genes identified by ChIP-sequencing. 293T cells stably expressing SFB-tagged FOXN2 or RFX1 were subjected to ChIP-sequencing using anti-FLAG antibody. Each experiment was performed with two biological replicates, and four control ChIP-sequencings were performed using 293T cells stably expressing other TFs.
- Reverse purification of FBXW11 (βTRCP2)-containing protein complexes conducted using the same TAP/MS protocol recovered FOXN2 as FBXW11-binding protein. Prey names, peptide counts and whether or not the interactions have been reported were listed.
- 293T cells were transfected with constructs encoding SFB-tagged FOXN2 and MYC-tagged βTRCP, its substrate binding-defective mutant βTRCP (R474A), or βTRCP2 as indicated. Pull-down experiments were carried out with S-protein beads and immunoblotted with antibodies as indicated.
- In vivo ubiquitination assays were performed by co-transfecting constructs encoding FLAG-tagged FOXN2, His-tagged ubiquitin, MYC-tagged βTRCP, βTRCP (R474A) or βTRCP2 into HEK293T cells as indicated. Cell lysates were denatured with 1% SDS and diluted 10-fold using PBS prior to the pull-down by Ni-NTA resin, followed by immunoblot with antibodies as indicated.
- 293T or 293T-shβTRCP2, 293T-shβTRCP2, 293T-shCUL1 cells were treated with 100 mM cycloheximide (CHX) for the indicated time. Immunoblotting was conducted with antibodies as indicated.
- A model showing on/off chromatin regulation of FOXN2 by transcriptional co-factors or E3 ligase complexes. All of the components indicated were identified from FOXN2 purifications.
Source data are available online for this figure.
Figure 6
Overview of JUN/CREB/ATF/NFATC1 subnetwork
- JUN/CREB/ATF/NFATC1 subnetwork map. Arrows indicate the identifications from TAP/MS. Bold arrows indicate the identifications from both TAP/MS and endogenous AP. Colours of arrows indicate the locations of interactions: red, in chromatin only; purple, in both fractions.
- NFATC1 binds to other factors mainly in chromatin fractions. 293T cells were transfected with constructs encoding MYC-tagged NFATC1 and SFB-tagged other TFs as indicated. Pull-down experiments were carried out with S-protein beads and immunoblotted with antibodies as indicated.
- ATF1 binds to ATF2, JUN and CREB1 in both fractions. 293T cells were transfected with constructs encoding MYC-tagged ATF1 and SFB-tagged other TFs as indicated. Pull-down experiments were carried out with S-protein beads and immunoblotted with antibodies as indicated.
- Overlap Venn diagram of CREB1, ATF2 and NFATC1 target genes using ChIP-seq data sets generated by the ENCODE consortium in GM12878 cells.
Source data are available online for this figure.
Comment in
- Changing partners: transcription factors form different complexes on and off chromatin.
Ji Z, Sharrocks AD. Ji Z, et al. Mol Syst Biol. 2015 Jan 21;11(1):782. doi: 10.15252/msb.20145936. Mol Syst Biol. 2015. PMID: 25609651 Free PMC article.
Similar articles
- Human transcription factor protein interaction networks.
Göös H, Kinnunen M, Salokas K, Tan Z, Liu X, Yadav L, Zhang Q, Wei GH, Varjosalo M. Göös H, et al. Nat Commun. 2022 Feb 9;13(1):766. doi: 10.1038/s41467-022-28341-5. Nat Commun. 2022. PMID: 35140242 Free PMC article. - Meta-Analysis of Transcriptome Regulation During Induction to Cardiac Myocyte Fate From Mouse and Human Fibroblasts.
Rastegar-Pouyani S, Khazaei N, Wee P, Yaqubi M, Mohammadnia A. Rastegar-Pouyani S, et al. J Cell Physiol. 2017 Aug;232(8):2053-2062. doi: 10.1002/jcp.25580. Epub 2017 Mar 24. J Cell Physiol. 2017. PMID: 27579918 - Analysis of transcriptional regulation of the small leucine rich proteoglycans.
Tasheva ES, Klocke B, Conrad GW. Tasheva ES, et al. Mol Vis. 2004 Oct 7;10:758-72. Mol Vis. 2004. PMID: 15496828 - Purification and characterization of transcription factors.
Nagore LI, Nadeau RJ, Guo Q, Jadhav YL, Jarrett HW, Haskins WE. Nagore LI, et al. Mass Spectrom Rev. 2013 Sep-Oct;32(5):386-98. doi: 10.1002/mas.21369. Epub 2013 Jul 7. Mass Spectrom Rev. 2013. PMID: 23832591 Free PMC article. Review. - Unravelling the biology of chromatin in health and cancer using proteomic approaches.
Eubanks CG, Dayebgadoh G, Liu X, Washburn MP. Eubanks CG, et al. Expert Rev Proteomics. 2017 Oct;14(10):905-915. doi: 10.1080/14789450.2017.1374860. Epub 2017 Sep 20. Expert Rev Proteomics. 2017. PMID: 28895440 Free PMC article. Review.
Cited by
- Unveiling the role of HP1α-HDAC1-STAT1 axis as a therapeutic target for HP1α-positive intrahepatic cholangiocarcinoma.
Xiong F, Wang D, Xiong W, Wang X, Huang WH, Wu GH, Liu WZ, Wang Q, Chen JS, Kuai YY, Wang B, Chen YJ. Xiong F, et al. J Exp Clin Cancer Res. 2024 May 30;43(1):152. doi: 10.1186/s13046-024-03070-3. J Exp Clin Cancer Res. 2024. PMID: 38812060 Free PMC article. - Radiosensitizing effect of curcumin-loaded lipid nanoparticles in breast cancer cells.
Minafra L, Porcino N, Bravatà V, Gaglio D, Bonanomi M, Amore E, Cammarata FP, Russo G, Militello C, Savoca G, Baglio M, Abbate B, Iacoviello G, Evangelista G, Gilardi MC, Bondì ML, Forte GI. Minafra L, et al. Sci Rep. 2019 Jul 31;9(1):11134. doi: 10.1038/s41598-019-47553-2. Sci Rep. 2019. PMID: 31366901 Free PMC article. - The Hippo Tumor Suppressor Pathway (YAP/TAZ/TEAD/MST/LATS) and EGFR-RAS-RAF-MEK in cancer metastasis.
Zinatizadeh MR, Miri SR, Zarandi PK, Chalbatani GM, Rapôso C, Mirzaei HR, Akbari ME, Mahmoodzadeh H. Zinatizadeh MR, et al. Genes Dis. 2019 Dec 5;8(1):48-60. doi: 10.1016/j.gendis.2019.11.003. eCollection 2021 Jan. Genes Dis. 2019. PMID: 33569513 Free PMC article. Review. - Regulation of TEAD Transcription Factors in Cancer Biology.
Huh HD, Kim DH, Jeong HS, Park HW. Huh HD, et al. Cells. 2019 Jun 17;8(6):600. doi: 10.3390/cells8060600. Cells. 2019. PMID: 31212916 Free PMC article. Review. - Unveiling the structure and interactions of SOG1, a NAC domain transcription factor: An in-silico perspective.
Mahapatra K. Mahapatra K. J Genet Eng Biotechnol. 2024 Mar;22(1):100333. doi: 10.1016/j.jgeb.2023.100333. Epub 2024 Jan 23. J Genet Eng Biotechnol. 2024. PMID: 38494249 Free PMC article.
References
- Altelaar AF, Munoz J, Heck AJ. Next-generation proteomics: towards an integrative view of proteome dynamics. Nat Rev Genet. 2013;14:35–48. - PubMed
- Baldwin AS., Jr The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- R01HG007538/HG/NHGRI NIH HHS/United States
- CA016672/CA/NCI NIH HHS/United States
- S10 OD012304/OD/NIH HHS/United States
- R01 HG007538/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous