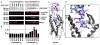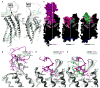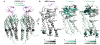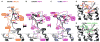Structural biology. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine - PubMed (original) (raw)
. 2015 Mar 6;347(6226):1117-22.
doi: 10.1126/science.1261064. Epub 2015 Jan 22.
Irina Kufareva 2, Lauren G Holden 1, Chong Wang 3, Yi Zheng 1, Chunxia Zhao 1, Gustavo Fenalti 3, Huixian Wu 3, Gye Won Han 4, Vadim Cherezov 5, Ruben Abagyan 1, Raymond C Stevens 6, Tracy M Handel 2
Affiliations
- PMID: 25612609
- PMCID: PMC4362693
- DOI: 10.1126/science.1261064
Structural biology. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine
Ling Qin et al. Science. 2015.
Abstract
Chemokines and their receptors control cell migration during development, immune system responses, and in numerous diseases, including inflammation and cancer. The structural basis of receptor:chemokine recognition has been a long-standing unanswered question due to the challenges of structure determination for membrane protein complexes. Here, we report the crystal structure of the chemokine receptor CXCR4 in complex with the viral chemokine antagonist vMIP-II at 3.1 angstrom resolution. The structure revealed a 1:1 stoichiometry and a more extensive binding interface than anticipated from the paradigmatic two-site model. The structure helped rationalize a large body of mutagenesis data and together with modeling provided insights into CXCR4 interactions with its endogenous ligand CXCL12, its ability to recognize diverse ligands, and the specificity of CC and CXC receptors for their respective chemokines.
Copyright © 2015, American Association for the Advancement of Science.
Figures
Fig. 1
Design and crystallization of a disulfide-trapped CXCR4:vMIP-II complex. (A) Non-reducing SDS-PAGE and Western blot of CXCR4(D97C) (left) and CXCR4(D187C) (right) coexpressed with cysteine mutants of vMIP-II (residues 1–7). Uncomplexed CXCR4 and disulfide-trapped complexes have molecular weights of approximately 45 kDa and 55 kDa, respectively. Band identities were confirmed by Western blot using antibodies against the FLAG and HA tags at the N- and C-termini of CXCR4 and vMIP-II, respectively (2nd and 3rd row). The 55 kDa band was labeled by anti-FLAG and anti-HA antibodies (2nd-4th row); the band at 45 kDa was only labeled by the anti-FLAG antibody (2nd and 4th row). (B) Thermal stability of the complexes measured by a CPM assay (40) are shown as mean ± SEM measurements performed in triplicate. (C) Overall structure of the CXCR4:vMIP-II complex (gray:magenta ribbon and transparent mesh). (D) Zoomed view of the vMIP-II N-terminus in the CXCR4 pocket showing the CXCR4(D187C):vMIP-II(W5C) disulfide bond. The 2mFo-DFc electron density map around the N-terminus is contoured at 1.0σ and colored blue.
Fig. 2
Interactions between CXCR4 and vMIP-II. (A–B) The interaction is mediated by a contiguous interface containing CRS1 (green), CRS2 (red) and CRS1.5 (blue). (A) The receptor is shown as a cut-open surface, the chemokine is shown as a ribbon, chemokine residues making substantial contacts with the receptor are shown as sticks. (B) The receptor is shown as a ribbon, receptor residues making substantial contacts with chemokine are shown as sticks, and vMIP-II is shown as a surface mesh. (C) Key residues (gray sticks) from CXCR4 (ribbon) that bind vMIP-II (surface representation). (D) Key residues (magenta sticks) from vMIP-II (white ribbon) that bind CXCR4 (cut-open surface). Non-carbon atoms are red (O), blue (N), and yellow (S); carbon stick color intensity is indicative of residue contact strength (Table S2).
Fig. 3
Comparison between CXCR4:vMIP-II and earlier CXCR4 structures. (A) Overlay of CXCR4 in the vMIP-II complex (gray), the IT1t complex (PDB ID 3ODU; cyan), and the CVX15 complex (PDB ID 3OE0; pale green). vMIP-II is shown as a gray transparent mesh. (B) CRS1 interaction between CXCR4 (gray) and vMIP-II (magenta), in comparison with IT1t-bound (cyan) and CVX15-bound (green) structures. Key residues mediating the CXCR4:vMIP-II interactions are shown as sticks. (C) Binding modes of vMIP-II, IT1t and CVX15 to CXCR4. CXCR4 is shown as a cut-open surface, colored by electrostatic potential; the bound ligands are shown as spheres. The white dotted line represents the boundary between the major and minor subpockets. (D–E) Comparison of CRS2 interactions of vMIP-II (magenta) with IT1t (cyan) and CVX15 (green).
Fig. 4
Crystallographic dimer of CXCR4:vMIP-II. (A) The overall dimer geometry is similar between CXCR4:vMIP-II (gray:magenta), CXCR4:IT1t (PDB ID 3ODU, light cyan:dark cyan), and CXCR4:CVX15 (PDB ID 3OE0, light green:green). (B) In all CXCR4 complexes solved thus far, the interaction between two CXCR4 molecules (ribbon) is mediated by the top halves of helix V, with additional contacts provided by either intracellular parts of helices III and V, or the top halves of helix VI (spheres). Views of the dimer interface on one monomer are shown for CXCR4:vMIP-II, CXCR4:IT1t, and CXCR4:CVX15 complexes. Residues that contribute to dimer formation are shown as spheres; color intensities represent contact strength.
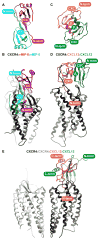
Fig. 5
Molecular models of homologous receptor:chemokine complexes: CXCR4:CXCL12 (A, D), CXCR4:vMIP-II (B), CCR5:vMIP-II (C), and CXCR4:CXCL12(P2G) (E). Panels A–C focus on CRS1 and CRS1.5 interactions while panels (D–E) show predicted CRS2 interactions. The dotted line indicates the approximate extracellular membrane-solvent boundary. (B) A model of WT CXCR4 sY21-F304 (gray) is built in complex with WT vMIP-II (magenta). Despite the absence of the D187C-W5C disulfide bond, the predicted interactions coincide precisely with those observed in the X-ray structure. vMIP-II W5 provides additional packing interaction with ECL2 of CXCR4. (A, C) Models of CXCR4:CXCL12 (gray:orange) and CCR5:vMIP-II (navy:magenta). (D–E) Models of CXCR4:CXCL12 (gray:orange) and CXCR4:CXCL12(P2G) (gray:green).
Fig. 6
Implications of the CXCR4:vMIP-II structure for understanding the stoichiometry of receptor:chemokine recognition. (A) vMIP-II (shown, PDB ID 2FHT) and other CC chemokine dimers are stabilized by β-sheet formation between the CC region and neighboring residues. (B) Superposition of the vMIP-II dimer onto the CXCR4:vMIP-II structure shows that binding of a CC dimer to the receptor is sterically impossible. (C) CXCL12 (shown, PDB ID 3GV3) and other CXC chemokine dimers are stabilized by β-sheet formation between their β1-strands. (D) Superposition of the CXCL12 dimer onto the CXCR4:vMIP-II structure shows that binding of a dimer is feasible. (E) If the receptor dimer geometry is relevant, and the vMIP-II orientation is predictive of CXC chemokine binding, CXC dimers do not simultaneously bind both receptors in a dimer.
Comment in
- Structural biology. Viral chemokine mimicry.
Standfuss J. Standfuss J. Science. 2015 Mar 6;347(6226):1071-2. doi: 10.1126/science.aaa7998. Science. 2015. PMID: 25745149 No abstract available.
Similar articles
- Structural basis for chemokine recognition by a G protein-coupled receptor and implications for receptor activation.
Ziarek JJ, Kleist AB, London N, Raveh B, Montpas N, Bonneterre J, St-Onge G, DiCosmo-Ponticello CJ, Koplinski CA, Roy I, Stephens B, Thelen S, Veldkamp CT, Coffman FD, Cohen MC, Dwinell MB, Thelen M, Peterson FC, Heveker N, Volkman BF. Ziarek JJ, et al. Sci Signal. 2017 Mar 21;10(471):eaah5756. doi: 10.1126/scisignal.aah5756. Sci Signal. 2017. PMID: 28325822 Free PMC article. - A novel peptide antagonist of CXCR4 derived from the N-terminus of viral chemokine vMIP-II.
Zhou N, Luo Z, Luo J, Hall JW, Huang Z. Zhou N, et al. Biochemistry. 2000 Apr 4;39(13):3782-7. doi: 10.1021/bi992750v. Biochemistry. 2000. PMID: 10736178 - Interaction of chemokine receptor CXCR4 in monomeric and dimeric state with its endogenous ligand CXCL12: coarse-grained simulations identify differences.
Cutolo P, Basdevant N, Bernadat G, Bachelerie F, Ha-Duong T. Cutolo P, et al. J Biomol Struct Dyn. 2017 Feb;35(2):399-412. doi: 10.1080/07391102.2016.1145142. Epub 2016 Jul 11. J Biomol Struct Dyn. 2017. PMID: 26813575 - Peptide and peptidomimetic ligands for CXC chemokine receptor 4 (CXCR4).
Oishi S, Fujii N. Oishi S, et al. Org Biomol Chem. 2012 Aug 14;10(30):5720-31. doi: 10.1039/c2ob25107h. Epub 2012 Apr 19. Org Biomol Chem. 2012. PMID: 22517031 Review. - Structural Analysis of Chemokine Receptor-Ligand Interactions.
Arimont M, Sun SL, Leurs R, Smit M, de Esch IJP, de Graaf C. Arimont M, et al. J Med Chem. 2017 Jun 22;60(12):4735-4779. doi: 10.1021/acs.jmedchem.6b01309. Epub 2017 Mar 10. J Med Chem. 2017. PMID: 28165741 Free PMC article. Review.
Cited by
- Post-expression strategies for structural investigations of membrane proteins.
Columbus L. Columbus L. Curr Opin Struct Biol. 2015 Jun;32:131-8. doi: 10.1016/j.sbi.2015.04.005. Epub 2015 May 16. Curr Opin Struct Biol. 2015. PMID: 25951412 Free PMC article. Review. - The chemokine X-factor: Structure-function analysis of the CXC motif at CXCR4 and ACKR3.
Wedemeyer MJ, Mahn SA, Getschman AE, Crawford KS, Peterson FC, Marchese A, McCorvy JD, Volkman BF. Wedemeyer MJ, et al. J Biol Chem. 2020 Oct 2;295(40):13927-13939. doi: 10.1074/jbc.RA120.014244. Epub 2020 Aug 11. J Biol Chem. 2020. PMID: 32788219 Free PMC article. - Interaction and dynamics of chemokine receptor CXCR4 binding with CXCL12 and hBD-3.
Penfield J, Zhang L. Penfield J, et al. Commun Chem. 2024 Sep 13;7(1):205. doi: 10.1038/s42004-024-01280-6. Commun Chem. 2024. PMID: 39271963 Free PMC article. - Modeling the complete chemokine-receptor interaction.
Wedemeyer MJ, Mueller BK, Bender BJ, Meiler J, Volkman BF. Wedemeyer MJ, et al. Methods Cell Biol. 2019;149:289-314. doi: 10.1016/bs.mcb.2018.09.005. Epub 2018 Nov 1. Methods Cell Biol. 2019. PMID: 30616825 Free PMC article. - Internal water channel formation in CXCR4 is crucial for Gi-protein coupling upon activation by CXCL12.
Chang CC, Liou JW, Dass KTP, Li YT, Jiang SJ, Pan SF, Yeh YC, Hsu HJ. Chang CC, et al. Commun Chem. 2020 Oct 8;3(1):133. doi: 10.1038/s42004-020-00383-0. Commun Chem. 2020. PMID: 36703316 Free PMC article.
References
- Nagasawa T, et al. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature. 1996;382:635. - PubMed
- Zou YR, et al. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 1998;393:595. - PubMed
- Koelink PJ, et al. Targeting chemokine receptors in chronic inflammatory diseases: An extensive review. Pharmacol Ther. 2012;133:1. - PubMed
- Balkwill F. The significance of cancer cell expression of the chemokine receptor CXCR4. Semin Cancer Biol. 2004;14:171. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM081763/GM/NIGMS NIH HHS/United States
- ACB-12002/PHS HHS/United States
- R21 AI101687/AI/NIAID NIH HHS/United States
- R01 GM117424/GM/NIGMS NIH HHS/United States
- R01 GM071872/GM/NIGMS NIH HHS/United States
- U01 GM094612/GM/NIGMS NIH HHS/United States
- AGM-12006/PHS HHS/United States
- U54 GM094618/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases