Collaborative computational project for electron cryo-microscopy - PubMed (original) (raw)
Collaborative computational project for electron cryo-microscopy
Chris Wood et al. Acta Crystallogr D Biol Crystallogr. 2015.
Abstract
The Collaborative Computational Project for Electron cryo-Microscopy (CCP-EM) has recently been established. The aims of the project are threefold: to build a coherent cryoEM community which will provide support for individual scientists and will act as a focal point for liaising with other communities, to support practising scientists in their use of cryoEM software and finally to support software developers in producing and disseminating robust and user-friendly programs. The project is closely modelled on CCP4 for macromolecular crystallography, and areas of common interest such as model fitting, underlying software libraries and tools for building program packages are being exploited. Nevertheless, cryoEM includes a number of techniques covering a large range of resolutions and a distinct project is required. In this article, progress so far is reported and future plans are discussed.
Keywords: CCP-EM; Collaborative Computational Project for Electron cryo-Microscopy.
Figures
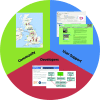
Figure 1
Overview of CCP-EM activities. The green sector refers to community-building activities, and is represented by a map of current groups performing high-resolution cryoEM. The blue sector refers to user-support activities, which are illustrated by the CCP-EM GUI currently under development and a snapshot of the CCP-EM website. Finally, the red sector refers to support of software developers, and is illustrated by the front page of CCPForge, which hosts the CCP-EM software project, and a schematic of the developing CCP-EM library.
Similar articles
- Recent developments in the CCP-EM software suite.
Burnley T, Palmer CM, Winn M. Burnley T, et al. Acta Crystallogr D Struct Biol. 2017 Jun 1;73(Pt 6):469-477. doi: 10.1107/S2059798317007859. Epub 2017 May 31. Acta Crystallogr D Struct Biol. 2017. PMID: 28580908 Free PMC article. - Current approaches for automated model building into cryo-EM maps using Buccaneer with CCP-EM.
Hoh SW, Burnley T, Cowtan K. Hoh SW, et al. Acta Crystallogr D Struct Biol. 2020 Jun 1;76(Pt 6):531-541. doi: 10.1107/S2059798320005513. Epub 2020 May 29. Acta Crystallogr D Struct Biol. 2020. PMID: 32496215 Free PMC article. - cryoem-cloud-tools: A software platform to deploy and manage cryo-EM jobs in the cloud.
Cianfrocco MA, Lahiri I, DiMaio F, Leschziner AE. Cianfrocco MA, et al. J Struct Biol. 2018 Sep;203(3):230-235. doi: 10.1016/j.jsb.2018.05.014. Epub 2018 Jun 1. J Struct Biol. 2018. PMID: 29864529 Free PMC article. - Progress in spatial resolution of structural analysis by cryo-EM.
Fukuda Y, Stapleton K, Kato T. Fukuda Y, et al. Microscopy (Oxf). 2023 Apr 6;72(2):135-143. doi: 10.1093/jmicro/dfac053. Microscopy (Oxf). 2023. PMID: 36269102 Review. - Sample preparation of biological macromolecular assemblies for the determination of high-resolution structures by cryo-electron microscopy.
Stark H, Chari A. Stark H, et al. Microscopy (Oxf). 2016 Feb;65(1):23-34. doi: 10.1093/jmicro/dfv367. Epub 2015 Dec 15. Microscopy (Oxf). 2016. PMID: 26671943 Review.
Cited by
- The cryo-EM structure of the endocytic receptor DEC-205.
Gully BS, Venugopal H, Fulcher AJ, Fu Z, Li J, Deuss FA, Llerena C, Heath WR, Lahoud MH, Caminschi I, Rossjohn J, Berry R. Gully BS, et al. J Biol Chem. 2021 Jan-Jun;296:100127. doi: 10.1074/jbc.RA120.016451. Epub 2020 Dec 3. J Biol Chem. 2021. PMID: 33257321 Free PMC article. - A national facility for biological cryo-electron microscopy.
Saibil HR, Grünewald K, Stuart DI. Saibil HR, et al. Acta Crystallogr D Biol Crystallogr. 2015 Jan 1;71(Pt 1):127-35. doi: 10.1107/S1399004714025280. Epub 2015 Jan 1. Acta Crystallogr D Biol Crystallogr. 2015. PMID: 25615867 Free PMC article. - Molecular mechanism of SbmA, a promiscuous transporter exploited by antimicrobial peptides.
Ghilarov D, Inaba-Inoue S, Stepien P, Qu F, Michalczyk E, Pakosz Z, Nomura N, Ogasawara S, Walker GC, Rebuffat S, Iwata S, Heddle JG, Beis K. Ghilarov D, et al. Sci Adv. 2021 Sep 10;7(37):eabj5363. doi: 10.1126/sciadv.abj5363. Epub 2021 Sep 8. Sci Adv. 2021. PMID: 34516884 Free PMC article. - A monoclonal antibody targeting a large surface of the receptor binding motif shows pan-neutralizing SARS-CoV-2 activity.
de Campos-Mata L, Trinité B, Modrego A, Tejedor Vaquero S, Pradenas E, Pons-Grífols A, Rodrigo Melero N, Carlero D, Marfil S, Santiago C, Raïch-Regué D, Bueno-Carrasco MT, Tarrés-Freixas F, Abancó F, Urrea V, Izquierdo-Useros N, Riveira-Muñoz E, Ballana E, Pérez M, Vergara-Alert J, Segalés J, Carolis C, Arranz R, Blanco J, Magri G. de Campos-Mata L, et al. Nat Commun. 2024 Feb 5;15(1):1051. doi: 10.1038/s41467-024-45171-9. Nat Commun. 2024. PMID: 38316751 Free PMC article. - X-Ray Crystallography for Macromolecular Complexes.
Fernández FJ, Querol-García J, Navas-Yuste S, Martino F, Vega MC. Fernández FJ, et al. Adv Exp Med Biol. 2024;3234:125-140. doi: 10.1007/978-3-031-52193-5_9. Adv Exp Med Biol. 2024. PMID: 38507204
References
- Crowther, R. A., Henderson, R. & Smith, J. M. (1996). J. Struct. Biol. 116, 9–16. - PubMed
Publication types
MeSH terms
Grants and funding
- MC_UP_A025_1013/MRC_/Medical Research Council/United Kingdom
- BB/G022577/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/L007835/1/MRC_/Medical Research Council/United Kingdom
- G0600084/MRC_/Medical Research Council/United Kingdom
- MR/J000825/1/Medical Research Council/United Kingdom
- R01 GM079429/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources