Disease-specific alterations in the enteric virome in inflammatory bowel disease - PubMed (original) (raw)
Multicenter Study
. 2015 Jan 29;160(3):447-60.
doi: 10.1016/j.cell.2015.01.002. Epub 2015 Jan 22.
Scott A Handley 1, Megan T Baldridge 1, Lindsay Droit 1, Catherine Y Liu 1, Brian C Keller 2, Amal Kambal 1, Cynthia L Monaco 2, Guoyan Zhao 3, Phillip Fleshner 4, Thaddeus S Stappenbeck 1, Dermot P B McGovern 5, Ali Keshavarzian 6, Ece A Mutlu 6, Jenny Sauk 7, Dirk Gevers 8, Ramnik J Xavier 9, David Wang 3, Miles Parkes 10, Herbert W Virgin 11
Affiliations
- PMID: 25619688
- PMCID: PMC4312520
- DOI: 10.1016/j.cell.2015.01.002
Multicenter Study
Disease-specific alterations in the enteric virome in inflammatory bowel disease
Jason M Norman et al. Cell. 2015.
Abstract
Decreases in the diversity of enteric bacterial populations are observed in patients with Crohn's disease (CD) and ulcerative colitis (UC). Less is known about the virome in these diseases. We show that the enteric virome is abnormal in CD and UC patients. In-depth analysis of preparations enriched for free virions in the intestine revealed that CD and UC were associated with a significant expansion of Caudovirales bacteriophages. The viromes of CD and UC patients were disease and cohort specific. Importantly, it did not appear that expansion and diversification of the enteric virome was secondary to changes in bacterial populations. These data support a model in which changes in the virome may contribute to intestinal inflammation and bacterial dysbiosis. We conclude that the virome is a candidate for contributing to, or being a biomarker for, human inflammatory bowel disease and speculate that the enteric virome may play a role in other diseases.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
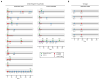
Figure 1. Virus Taxonomic Assignment and Imbalance in IBD
A) Relative abundance of sequences assigned to the indicated viral taxa. Error bars represent the mean +/− SD. B) Correlation plot of the Caudovirales and Microviridae relative abundance for all samples. Linear regression +/− 95% confidence interval and Spearman correlation coefficient are shown. C) Microviridae and Caudovirales relative abundance for United Kingdom household controls, UC and CD (top); Los Angeles and Chicago IBD (bottom). The bars indicate the median and interquartile range. Statistical significance was determined by the Mann-Whitney test. See also Tables S1, S2, and S3.
Figure 2. In-depth, Longitudinal Cohort Graphical Timeline
A) United Kingdom IBD stool samples and non-IBD household control stools were collected at the onset of symptoms (flare) and collected as symptoms resolved where indicated. The length of each IBD flare is indicated by the red shaded oval. The samples are annotated by household ID and sample number. B) Chicago UC samples were collected first during inactive disease and again when symptoms exacerbated. The samples are annotated by the subject and sample number. See also Tables S1 and S2.
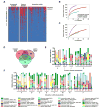
Figure 3. Bacteriophage Expansion is Associated with CD and UC
A) Presence-absence heat map of the sequences assigned to Caudovirales taxa in VLP preparations from UK household control, CD and UC stool samples. B and C) Rarefaction curves of Caudovirales richness versus an increasing number of sub-samplings with replacement. B) Caudovirales richness based on individual sequences. C) Richness based on assembled Caudovirales contigs. The curves represent the average of 500 iterations at each depth of samples. D) Venn diagram of the Caudovirales taxa in household control, CD and UC samples. N = the number of samples within each sub-group. E and F) Plots of the relative abundance of the 35 most abundant Caudovirales taxa in the UK UC and CD households. Bars are annotated by the household ID and sample number. Green numbers = household controls; purple numbers = CD; red numbers = UC. See also Figure S1, Tables S4, S5, and S6.
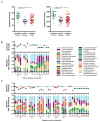
Figure 4. Alterations in the Bacterial Community Composition in IBD
A) Alpha diversity (left) and bacterial species richness (right) based on 16S rRNA gene sequences in the stool of household controls, CD and UC patients. Statistical significance was determined by the Kruskal-Wallis test with Dunn’s correction comparing all samples to all samples. ** = p > 0.01. B and C) Plots of Alpha diversity normalized to the diversity in household controls (top) and relative bacterial family abundance (bottom) of UK B) CD households and C) UC households. Green numbers = household controls; purple numbers = CD; red numbers = UC. See also Tables S5, S7, and S8.
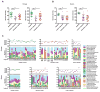
Figure 5. Disease-Specific Bacteria-Caudovirales Patterns in IBD
A) Spearman correlation plot of Caudovirales richness, Caudovirales Shannon diversity, bacterial alpha diversity, and bacterial species richness for UK household control, CD, and UC samples. Statistical significance was determined for all pair-wise comparisons; those with p values < 0.05 are indicated. Positive values (blue circles) indicate positive correlations and negative values (red circles) indicate inverse correlations. The size and shading of the circle indicates the magnitude of the correlation where darker shades are more correlated than lighter shades. B) Spearman correlation plots of the relative abundances of the 50 most abundant Caudovirales taxa and bacterial families identified to be significantly associated with disease. UK household control (top), CD (middle), and UC (bottom) samples. The gray bars indicate any taxa that were not detected in the cohort sub-group. Statistical significance was determined for all pair-wise comparisons; only significant (p value < 0.05) are displayed. See also Figure S2.
Figure 6. Validation of Bacterial Dysbiosis in Two Additional Cohorts
A and B) Faith’s phylogenetic alpha diversity (left) and bacterial species richness (right) based on 16S rRNA gene sequences in the stool of A) Chicago healthy controls, CD, and UC patients and B) Boston healthy controls, CD, and UC patients. Statistical significance was determined by the Kruskal-Wallis test with Dunn’s correction comparing all samples to all samples. * = p > 0.05, ** = p > 0.01. C) Plot of alpha diversity normalized to the average diversity in healthy subjects (top) and relative bacterial family abundance (bottom) for the Chicago and Boston cohorts. Healthy control, UC, and CD samples are indicated. Longitudinal samples from the same subject are grouped together. See also Tables S5, S7, and S8.
Figure 7. Validation of Caudovirales Expansion in Two Additional Clinical Cohorts
A and B) Rarefaction curves of Caudovirales richness versus an increasing number of sub-samplings with replacement for the Chicago (top) and Boston (bottom) cohorts. A) Caudovirales richness based on individual sequences. B) Richness based on Caudovirales contigs. C) Venn diagram of the Caudovirales taxa in healthy control, CD and UC samples from Chicago (top) and Boston (bottom). N = the number of samples within each sub-group. See also Tables S4, S5, and S6.
Comment in
- IBD. Gut microbiota in IBD goes viral.
Ray K. Ray K. Nat Rev Gastroenterol Hepatol. 2015 Mar;12(3):122. doi: 10.1038/nrgastro.2015.26. Epub 2015 Feb 10. Nat Rev Gastroenterol Hepatol. 2015. PMID: 25666643 No abstract available. - The Enteric Virome in Inflammatory Bowel Disease.
Brooks J, Watson AJ. Brooks J, et al. Gastroenterology. 2015 Oct;149(4):1120-1. doi: 10.1053/j.gastro.2015.08.022. Epub 2015 Aug 25. Gastroenterology. 2015. PMID: 26319032 Free PMC article. No abstract available.
Similar articles
- The Enteric Virome in Inflammatory Bowel Disease.
Brooks J, Watson AJ. Brooks J, et al. Gastroenterology. 2015 Oct;149(4):1120-1. doi: 10.1053/j.gastro.2015.08.022. Epub 2015 Aug 25. Gastroenterology. 2015. PMID: 26319032 Free PMC article. No abstract available. - IBD. Gut microbiota in IBD goes viral.
Ray K. Ray K. Nat Rev Gastroenterol Hepatol. 2015 Mar;12(3):122. doi: 10.1038/nrgastro.2015.26. Epub 2015 Feb 10. Nat Rev Gastroenterol Hepatol. 2015. PMID: 25666643 No abstract available. - Gut mucosal virome alterations in ulcerative colitis.
Zuo T, Lu XJ, Zhang Y, Cheung CP, Lam S, Zhang F, Tang W, Ching JYL, Zhao R, Chan PKS, Sung JJY, Yu J, Chan FKL, Cao Q, Sheng JQ, Ng SC. Zuo T, et al. Gut. 2019 Jul;68(7):1169-1179. doi: 10.1136/gutjnl-2018-318131. Epub 2019 Mar 6. Gut. 2019. PMID: 30842211 Free PMC article. - Enteric microbiota leads to new therapeutic strategies for ulcerative colitis.
Chen WX, Ren LH, Shi RH. Chen WX, et al. World J Gastroenterol. 2014 Nov 14;20(42):15657-63. doi: 10.3748/wjg.v20.i42.15657. World J Gastroenterol. 2014. PMID: 25400449 Free PMC article. Review. - Mucosal barrier, bacteria and inflammatory bowel disease: possibilities for therapy.
Merga Y, Campbell BJ, Rhodes JM. Merga Y, et al. Dig Dis. 2014;32(4):475-83. doi: 10.1159/000358156. Epub 2014 Jun 23. Dig Dis. 2014. PMID: 24969297 Review.
Cited by
- Hidden players: the bacteria-killing viruses of the gut microbiome.
King A. King A. Nature. 2024 Oct 31. doi: 10.1038/d41586-024-03532-w. Online ahead of print. Nature. 2024. PMID: 39482427 No abstract available. - A single-stranded based library preparation method for virome characterization.
Zhai X, Gobbi A, Kot W, Krych L, Nielsen DS, Deng L. Zhai X, et al. Microbiome. 2024 Oct 24;12(1):219. doi: 10.1186/s40168-024-01935-5. Microbiome. 2024. PMID: 39449043 Free PMC article. - Host-pathobiont interactions in Crohn's disease.
Caruso R, Lo BC, Chen GY, Núñez G. Caruso R, et al. Nat Rev Gastroenterol Hepatol. 2024 Oct 24. doi: 10.1038/s41575-024-00997-y. Online ahead of print. Nat Rev Gastroenterol Hepatol. 2024. PMID: 39448837 Review. - Alterations of the gut microbiome in HIV infection highlight human anelloviruses as potential predictors of immune recovery.
Boukadida C, Peralta-Prado A, Chávez-Torres M, Romero-Mora K, Rincon-Rubio A, Ávila-Ríos S, Garrido-Rodríguez D, Reyes-Terán G, Pinto-Cardoso S. Boukadida C, et al. Microbiome. 2024 Oct 17;12(1):204. doi: 10.1186/s40168-024-01925-7. Microbiome. 2024. PMID: 39420423 Free PMC article. - Growth stage-related capsular polysaccharide translocon Wza in Vibrio splendidus modifies phage vB_VspM_VS2 susceptibility.
Jiang L, Wen J, Tan D, Xie J, Li J, Li C. Jiang L, et al. Commun Biol. 2024 Oct 16;7(1):1338. doi: 10.1038/s42003-024-07038-z. Commun Biol. 2024. PMID: 39414953 Free PMC article.
References
- Angelberger S, Reinisch W, Makristathis A, Lichtenberger C, Dejaco C, Papay P, Novacek G, Trauner M, Loy A, Berry D. Temporal bacterial community dynamics vary among ulcerative colitis patients after fecal microbiota transplantation. The American Journal of Gastroenterology. 2013;108:1620–1630. - PubMed
- Basic M, Keubler LM, Buettner M, Achard M, Breves G, Schroder B, Smoczek A, Jorns A, Wedekind D, Zschemisch NH, et al. Norovirus Triggered Microbiota-driven Mucosal Inflammation in Interleukin 10-deficient Mice. Inflammatory bowel diseases. 2014;20:431–443. - PubMed
Publication types
MeSH terms
Grants and funding
- U54DE023789-01/DE/NIDCR NIH HHS/United States
- R01 AI084887/AI/NIAID NIH HHS/United States
- AT001628/AT/NCCIH NIH HHS/United States
- U54 DE023789/DE/NIDCR NIH HHS/United States
- DK046763-19/DK/NIDDK NIH HHS/United States
- U01 DK062413/DK/NIDDK NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- 5T32AI007172-34/AI/NIAID NIH HHS/United States
- R01 DK092405/DK/NIDDK NIH HHS/United States
- Department of Health/United Kingdom
- T32 HL007317/HL/NHLBI NIH HHS/United States
- T32 CA009547/CA/NCI NIH HHS/United States
- P30 CA91842/CA/NCI NIH HHS/United States
- R01 HS021747/HS/AHRQ HHS/United States
- R01 AT007143/AT/NCCIH NIH HHS/United States
- 5T32CA009547/CA/NCI NIH HHS/United States
- U54 DE023798/DE/NIDCR NIH HHS/United States
- T32 AI007163/AI/NIAID NIH HHS/United States
- UL1TR000448/TR/NCATS NIH HHS/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- T32 AI007172/AI/NIAID NIH HHS/United States
- R21 DK071838/DK/NIDDK NIH HHS/United States
- UL1 TR000448/TR/NCATS NIH HHS/United States
- AI067068/AI/NIAID NIH HHS/United States
- U54 AI057160/AI/NIAID NIH HHS/United States
- 5T32AI007163-35/AI/NIAID NIH HHS/United States
- HS021747/HS/AHRQ HHS/United States
- R01 OD011170/OD/NIH HHS/United States
- T32 HL007317-36/HL/NHLBI NIH HHS/United States
- U01 AI067068/AI/NIAID NIH HHS/United States
- P01 DK046763/DK/NIDDK NIH HHS/United States
- DK071838/DK/NIDDK NIH HHS/United States
- DK062413/DK/NIDDK NIH HHS/United States
- R21 AT001628/AT/NCCIH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical