New genetic variants of LATS1 detected in urinary bladder and colon cancer - PubMed (original) (raw)
New genetic variants of LATS1 detected in urinary bladder and colon cancer
Mona K Saadeldin et al. Front Genet. 2015.
Abstract
LATS1, the large tumor suppressor 1 gene, encodes for a serine/threonine kinase protein and is implicated in cell cycle progression. LATS1 is down-regulated in various human cancers, such as breast cancer, and astrocytoma. Point mutations in LATS1 were reported in human sarcomas. Additionally, loss of heterozygosity of LATS1 chromosomal region predisposes to breast, ovarian, and cervical tumors. In the current study, we investigated LATS1 genetic variations including single nucleotide polymorphisms (SNPs), in 28 Egyptian patients with either urinary bladder or colon cancers. The LATS1 gene was amplified and sequenced and the expression of LATS1 at the RNA level was assessed in 12 urinary bladder cancer samples. We report, the identification of a total of 29 variants including previously identified SNPs within LATS1 coding and non-coding sequences. A total of 18 variants were novel. Majority of the novel variants, 13, were mapped to intronic sequences and un-translated regions of the gene. Four of the five novel variants located in the coding region of the gene, represented missense mutations within the serine/threonine kinase catalytic domain. Interestingly, LATS1 RNA steady state levels was lost in urinary bladder cancerous tissue harboring four specific SNPs (16045 + 41736 + 34614 + 56177) positioned in the 5'UTR, intron 6, and two silent mutations within exon 4 and exon 8, respectively. This study identifies novel single-base-sequence alterations in the LATS1 gene. These newly identified variants could potentially be used as novel diagnostic or prognostic tools in cancer.
Keywords: LATS1; colon cancer; genetic variants; tumor suppressor; urinary bladder cancer.
Figures
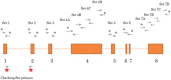
Figure 1
Schematic representation of primers sets used to amplify the LATS1 gene. 13 sets of primers were designed (sequence details shown in Supplemental Table 1) to amplify and sequence LATS1. Primer sets with an asterisk are the sets used for amplification and sequencing of the LATS1 8 exon and the adjacent 5′ and 3′ intronic region. The sets without an asterisk are used in the sequencing only. The primer sets used to assess LATS1 expression are presented with the red stars.
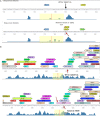
Figure 2
LATS1 expressions analysis using PCR. LATS1 expression was examined in urinary bladder cancer tissues (UC), peritumoral urinary bladder (UN). UC6 did not contain any SNP in the LATS1 gene. UC7 showed SNP at 41736; UC14 harbors SNP at 35366; UC1 has SNPs/variants at 16045, 34614, 35366, 41625, 41736, and 56177; UC4 has SNPs/variants at 34614, 35366, 41625, and 41736; UC5 has SNPs/variants at 34614, 41625, 41736, and 42029; UC8 has SNPs/variants at 35366, 41625, 41736, 42029, and 56177; UC11 has SNPs at 16045, 34614, 41736, and 56177; UC13 harbors SNPs/variants at 16045, 35366, 41736, and 42029; UC 15 harbors SNPs/variants at 16045, 34614, 35366, 41736, 42029, and 56177; UC 20 harbors SNPs at 16045, 35366, and 41736. UN3 harbors SNPs/variants at 515, 15943, 41736, 56177, 56152, and 56164. GAPDH was used as an endogenous control. Generuler 1 Kb Plus DNA Ladder (Fermentas®) was used as a size marker.
Figure 3
Schematic diagram of the Transfac predicted TF binding sites within a fragment of LATS1 gene. (A) Transfac diagram of the wild type (WT) fragment of LATS1 intron-1 that depicts the myogenin transcription factor-binding site as created by the variant at consensus position 15943. (B) Diagram of the depicted SREBP TF binding site that is generated by the variant at 42029 (indicated by an arrow) at LATS1 intron-6 as well as the predicted TF binding sites that were interrupted by the variant (Kel et al., ; Matys et al., 2006).
Similar articles
- Contribution of germline BRCA1 and BRCA2 sequence alterations to breast cancer in Northern India.
Saxena S, Chakraborty A, Kaushal M, Kotwal S, Bhatanager D, Mohil RS, Chintamani C, Aggarwal AK, Sharma VK, Sharma PC, Lenoir G, Goldgar DE, Szabo CI. Saxena S, et al. BMC Med Genet. 2006 Oct 4;7:75. doi: 10.1186/1471-2350-7-75. BMC Med Genet. 2006. PMID: 17018160 Free PMC article. - Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers.
Takahashi Y, Miyoshi Y, Takahata C, Irahara N, Taguchi T, Tamaki Y, Noguchi S. Takahashi Y, et al. Clin Cancer Res. 2005 Feb 15;11(4):1380-5. doi: 10.1158/1078-0432.CCR-04-1773. Clin Cancer Res. 2005. PMID: 15746036 - Molecular alterations of h-warts/LATS1 tumor suppressor in human soft tissue sarcoma.
Hisaoka M, Tanaka A, Hashimoto H. Hisaoka M, et al. Lab Invest. 2002 Oct;82(10):1427-35. doi: 10.1097/01.lab.0000032381.68634.ca. Lab Invest. 2002. PMID: 12379777 - Hypermethylation of promoter region of LATS1--a CDK interacting protein in oral squamous cell carcinomas--a pilot study in India.
Reddy VR, Annamalai T, Narayanan V, Ramanathan A. Reddy VR, et al. Asian Pac J Cancer Prev. 2015;16(4):1599-603. doi: 10.7314/apjcp.2015.16.4.1599. Asian Pac J Cancer Prev. 2015. PMID: 25743838 - Genetic variants in urinary bladder cancer: collective power of the "wimp SNPs".
Golka K, Selinski S, Lehmann ML, Blaszkewicz M, Marchan R, Ickstadt K, Schwender H, Bolt HM, Hengstler JG. Golka K, et al. Arch Toxicol. 2011 Jun;85(6):539-54. doi: 10.1007/s00204-011-0676-3. Epub 2011 Mar 5. Arch Toxicol. 2011. PMID: 21380501 Review.
Cited by
- YAP/TAZ Activation as a Target for Treating Metastatic Cancer.
Warren JSA, Xiao Y, Lamar JM. Warren JSA, et al. Cancers (Basel). 2018 Apr 10;10(4):115. doi: 10.3390/cancers10040115. Cancers (Basel). 2018. PMID: 29642615 Free PMC article. Review. - Dendritic cell vaccine immunotherapy; the beginning of the end of cancer and COVID-19. A hypothesis.
Saadeldin MK, Abdel-Aziz AK, Abdellatif A. Saadeldin MK, et al. Med Hypotheses. 2021 Jan;146:110365. doi: 10.1016/j.mehy.2020.110365. Epub 2020 Nov 9. Med Hypotheses. 2021. PMID: 33221134 Free PMC article. Review. - Loss of large tumor suppressor 1 promotes growth and metastasis of gastric cancer cells through upregulation of the YAP signaling.
Zhang J, Wang G, Chu SJ, Zhu JS, Zhang R, Lu WW, Xia LQ, Lu YM, Da W, Sun Q. Zhang J, et al. Oncotarget. 2016 Mar 29;7(13):16180-93. doi: 10.18632/oncotarget.7568. Oncotarget. 2016. PMID: 26921249 Free PMC article. - Hippo signaling pathway in companion animal diseases, an under investigated signaling cascade.
Budel SJ, Penning MM, Penning LC. Budel SJ, et al. Vet Q. 2021 Dec;41(1):172-180. doi: 10.1080/01652176.2021.1923085. Vet Q. 2021. PMID: 33945400 Free PMC article. - The Hippo pathway: an emerging role in urologic cancers.
Cinar B, Alp E, Al-Mathkour M, Boston A, Dwead A, Khazaw K, Gregory A. Cinar B, et al. Am J Clin Exp Urol. 2021 Aug 25;9(4):301-317. eCollection 2021. Am J Clin Exp Urol. 2021. PMID: 34541029 Free PMC article. Review.
References
- Hisaoka M., Tanaka A., Hashimoto H. (2002). Molecular alterations of h-warts/LATS1 tumor suppressor in human soft tissue sarcoma. Lab. Invest. 82, 1427–1435. 10.1097/01.LAB.0000032381.68634.CA - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources