Human promoters are intrinsically directional - PubMed (original) (raw)
Human promoters are intrinsically directional
Sascha H C Duttke et al. Mol Cell. 2015.
Abstract
Divergent transcription, in which reverse-oriented transcripts occur upstream of eukaryotic promoters in regions devoid of annotated genes, has been suggested to be a general property of active promoters. Here we show that the human basal RNA polymerase II transcriptional machinery and core promoter are inherently unidirectional and that reverse-oriented transcripts originate from their own cognate reverse-directed core promoters. In vitro transcription analysis and mapping of nascent transcripts in HeLa cells revealed that sequences at reverse start sites are similar to those of their forward counterparts. The use of DNase I accessibility to define proximal promoter borders revealed that about half of promoters are unidirectional and that unidirectional promoters are depleted at their upstream edges of reverse core promoter sequences and their associated chromatin features. Divergent transcription is thus not an inherent property of the transcription process but rather the consequence of the presence of both forward- and reverse-directed core promoters.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
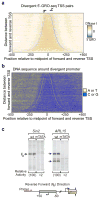
Figure 1. Directional transcription of core promoters
a, Unidirectional transcription of diverse types of core promoters, ± 50 bp in respect to the +1 TSS (marked by the arrow) in vitro. See also Figure S1. b, Directionality of the core promoter and promoter regions (n = 15,474) as mapped by 5′-GRO-Seq in HeLa S3 cells, plotted as percent antisense activity (5′end read counts in a given antisense window divided by that number plus the counts in the forward TSS cluster) for different windows upstream of forward TSS (see Experimental Procedures). Blue = −50 to +50, orange = −150 to −50, red = −250 to −150, and green = −350 to −250. c, Distribution of distances between divergent pairs of 5′GRO-Seq-defined TSSs (n = 3865). Reverse transcription start sites were mapped relative to their corresponding +1 forward start site.
Figure 2. Transcription initiation from divergent core promoters occurs at edges of open chromatin
a, Normalized 5′-GRO-Seq (blue) read 5′end counts and DNaseI-seq (orange) read 5′end counts in bins of 10, ±0.5 kb from the center point of divergent TSS pairs (n = 3865; see Experimental Procedures), ranked from top to bottom by increasing distance between pairs. b, Genomic DNA sequence of divergent TSS pairs centered and ranked as in “a”. Bases ‘A’ and ‘T’ are yellow, bases ‘C’ and ‘G’ are blue. See also Figures S2 and S3. c, TATA-sensitive in vitro transcription of reverse directed core promoters. +1 TSS is marked by the arrow.
Figure 3. Examples of divergent, unidirectional, and bidirectional transcription
Browser snapshots of examples displaying genes where divergent transcription is absent (a; unidirectional), present (b; divergent), or occurring at annotated bidirectional genes (c; bidirectional). Shown is DNaseI-seq signal as generated by JAMM in black and 5′GRO-seq reads in red for the + strand and blue for the − strand.
Figure 4. Many promoter DHSs lack core promoter sequences necessary for divergent transcription
a, b, Normalized 5′-GRO-Seq (blue) read 5′end counts and DNaseI-seq (orange) read 5′end counts in bins of 10 bp, ±1 kb from the center point of divergent (a; n = 1741) and unidirectional (b; n = 2237) promoter DHSs (see Experimental Procedures) ranked from top to bottom by increasing DHS width. See also Figure S4. c, Normalized, fragment-extended H2AZ (top) and TAF1 (bottom) ChIP-seq read counts in bins of 10 bp for divergent (left) and unidirectional (right) DHSs centered and ranked as above. See also Figure S5. d, Predicted TSS scores around corresponding DHS edges resulting from a position-specific Markov chain model (see Experimental Procedures) trained on ±50 nucleotides around divergent forward TSS. Blue = divergent forward, light blue = divergent reverse, red = unidirectional forward, orange = unidirectional reverse. See also Tables S1 and S2.
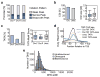
Figure 5. Characteristics of divergent and unidirectional promoters
a, Percentage of initiation patterns defined by Ni et al. for forward 5′-GRO-seq clusters of the DHS-defined divergent and unidirectional promoter groups and reverse clusters of divergent group. b, Percentage of divergent or unidirectional promoter DHSs intersecting an annotated CpG island (left; see Experimental Procedures). Size distributions of CpG islands that intersect divergent or unidirectional promoter DHSs (right; see Experimental Procedures). c, Percent of forward direction 5′-GRO-seq clusters containing a TATA motif match from −35 to −25 relative to the cluster mode for forward 5′-GRO-seq clusters of divergent and unidirectional promoters and reverse clusters of the divergent group (left; see Experimental Procedures) and the distributions of the corresponding scores (right). d, Positional average fragment-extended ChIP-seq read counts within TBP peak summits as called by SISSRS in bins of 10 nucleotides for TATA-containing and TATA-less forward and reverse core promoter subsets of divergent or forward only for unidirectional promoters (see Experimental Procedures). e, Distributions of DHS widths for unidirectional, divergent, and bidirectional promoter groups. See also Figures S4 and S5 and Table S2.
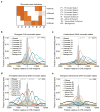
Figure 6. Distinct chromatin environment at unidirectional, bidirectional, and divergent promoter DHSs
a, Chromatin state definitions based on Hidden Markov Model clustering of histone modification ChIP-Seq signal at 10 base pair resolution (see Experimental Procedures). Each state is a multivariate Gaussian distribution. Shown are the distribution mean vectors representing scaled, normalized ChIP-Seq signal. b, c, d, e, Chromatin state coverage ±2kb around the center of divergent promoter DHS (b), unidirectional promoter DHS (c), bidirectional promoter DHS (d), and divergent intergenic DHS (e) at single nucleotide resolution. Grey = DNaseI-seq read 5′end counts, red = Promoter State1, blue = Promoter State 2, green = Promoter State 3, light blue = Promoter State 4, black = Inactive Enhancer, yellow = Active Enhancer, pink = Transcribed Enhancer, orange = Background. See also Figure S6.
Figure 7. Model of divergent, bidirectional, and unidirectional promoters
For each type of promoter, +1 and −1 nucleosome positions occur at variable spacing from each other, forward gene transcription initiates just inside the downstream edge of the NFR, and the +1 nucleosome is modified with H3K4me3 and H3K27ac. When transcription initiation occurs from the upstream NFR edge on the opposite strand from the forward gene, the −1 nucleosome gets similarly modified when stable, annotated transcripts are present (for bidirectional promoters), or is enriched for H3K4me2, in addition to H3K4me3 and H3K27ac, when divergent transcription occurs (i.e., when unstable non-coding transcripts are generated).
Comment in
- Human Gene Promoters Are Intrinsically Bidirectional.
Andersson R, Chen Y, Core L, Lis JT, Sandelin A, Jensen TH. Andersson R, et al. Mol Cell. 2015 Nov 5;60(3):346-7. doi: 10.1016/j.molcel.2015.10.015. Mol Cell. 2015. PMID: 26545074 Free PMC article. No abstract available. - Perspectives on Unidirectional versus Divergent Transcription.
Duttke SH, Lacadie SA, Ibrahim MM, Glass CK, Corcoran DL, Benner C, Heinz S, Kadonaga JT, Ohler U. Duttke SH, et al. Mol Cell. 2015 Nov 5;60(3):348-9. doi: 10.1016/j.molcel.2015.10.014. Mol Cell. 2015. PMID: 26545075 Free PMC article. No abstract available.
Similar articles
- RNA exosome depletion reveals transcription upstream of active human promoters.
Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. Preker P, et al. Science. 2008 Dec 19;322(5909):1851-4. doi: 10.1126/science.1164096. Epub 2008 Dec 4. Science. 2008. PMID: 19056938 - Eukaryotic core promoters and the functional basis of transcription initiation.
Haberle V, Stark A. Haberle V, et al. Nat Rev Mol Cell Biol. 2018 Oct;19(10):621-637. doi: 10.1038/s41580-018-0028-8. Nat Rev Mol Cell Biol. 2018. PMID: 29946135 Free PMC article. Review. - Structural features of DNA that determine RNA polymerase II core promoter.
Il'icheva IA, Khodikov MV, Poptsova MS, Nechipurenko DY, Nechipurenko YD, Grokhovsky SL. Il'icheva IA, et al. BMC Genomics. 2016 Nov 25;17(1):973. doi: 10.1186/s12864-016-3292-z. BMC Genomics. 2016. PMID: 27884105 Free PMC article. - Bidirectional promoters in the transcription of mammalian genomes.
Orekhova AS, Rubtsov PM. Orekhova AS, et al. Biochemistry (Mosc). 2013 Apr;78(4):335-41. doi: 10.1134/S0006297913040020. Biochemistry (Mosc). 2013. PMID: 23590436 Review.
Cited by
- Differential regulation of CpG island methylation within divergent and unidirectional promoters in colorectal cancer.
Namba S, Sato K, Kojima S, Ueno T, Yamamoto Y, Tanaka Y, Inoue S, Nagae G, Iinuma H, Hazama S, Ishihara S, Aburatani H, Mano H, Kawazu M. Namba S, et al. Cancer Sci. 2019 Mar;110(3):1096-1104. doi: 10.1111/cas.13937. Epub 2019 Mar 2. Cancer Sci. 2019. PMID: 30637877 Free PMC article. - Widespread Transcriptional Scanning in the Testis Modulates Gene Evolution Rates.
Xia B, Yan Y, Baron M, Wagner F, Barkley D, Chiodin M, Kim SY, Keefe DL, Alukal JP, Boeke JD, Yanai I. Xia B, et al. Cell. 2020 Jan 23;180(2):248-262.e21. doi: 10.1016/j.cell.2019.12.015. Cell. 2020. PMID: 31978344 Free PMC article. - In the loop: promoter-enhancer interactions and bioinformatics.
Mora A, Sandve GK, Gabrielsen OS, Eskeland R. Mora A, et al. Brief Bioinform. 2016 Nov;17(6):980-995. doi: 10.1093/bib/bbv097. Epub 2015 Nov 19. Brief Bioinform. 2016. PMID: 26586731 Free PMC article. Review. - Core promoter-specific gene regulation: TATA box selectivity and Initiator-dependent bi-directionality of serum response factor-activated transcription.
Xu M, Gonzalez-Hurtado E, Martinez E. Xu M, et al. Biochim Biophys Acta. 2016 Apr;1859(4):553-63. doi: 10.1016/j.bbagrm.2016.01.005. Epub 2016 Jan 26. Biochim Biophys Acta. 2016. PMID: 26824723 Free PMC article. - Single-molecule nascent RNA sequencing identifies regulatory domain architecture at promoters and enhancers.
Tome JM, Tippens ND, Lis JT. Tome JM, et al. Nat Genet. 2018 Nov;50(11):1533-1541. doi: 10.1038/s41588-018-0234-5. Epub 2018 Oct 22. Nat Genet. 2018. PMID: 30349116 Free PMC article.
References
- Adachi N, Lieber MR. Bidirectional gene organization: a common architectural feature of the human genome. Cell. 2002;109:807–809. - PubMed
- Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Molecular Cell. 2006;23:841–851. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases