Complete plastome sequence of Thalictrum coreanum (Ranunculaceae) and transfer of the rpl32 gene to the nucleus in the ancestor of the subfamily Thalictroideae - PubMed (original) (raw)
Complete plastome sequence of Thalictrum coreanum (Ranunculaceae) and transfer of the rpl32 gene to the nucleus in the ancestor of the subfamily Thalictroideae
Seongjun Park et al. BMC Plant Biol. 2015.
Abstract
Background: Plastids originated from cyanobacteria and the majority of the ancestral genes were lost or functionally transferred to the nucleus after endosymbiosis. Comparative genomic investigations have shown that gene transfer from plastids to the nucleus is an ongoing evolutionary process but molecular evidence for recent functional gene transfers among seed plants have only been documented for the four genes accD, infA, rpl22, and rpl32.
Results: The complete plastid genome of Thalictrum coreanum, the first from the subfamily Thalictroideae (Ranunculaceae), was sequenced and revealed the losses of two genes, infA and rpl32. The functional transfer of these two genes to the nucleus in Thalictrum was verified by examination of nuclear transcriptomes. A survey of the phylogenetic distribution of the rpl32 loss was performed using 17 species of Thalictrum and representatives of related genera in the subfamily Thalictroideae. The plastid-encoded rpl32 gene is likely nonfunctional in members of the subfamily Thalictroideae (Aquilegia, Enemion, Isopyrum, Leptopyrum, Paraquilegia, and Semiaquilegia) including 17 Thalictrum species due to the presence of indels that disrupt the reading frame. A nuclear-encoded rpl32 with high sequence identity was identified in both Thalictrum and Aquilegia. The phylogenetic distribution of this gene loss/transfer and the high level of sequence similarity in transit peptides suggest a single transfer of the plastid-encoded rpl32 to the nucleus in the ancestor of the subfamily Thalictroideae approximately 20-32 Mya.
Conclusions: The genome sequence of Thalictrum coreanum provides valuable information for improving the understanding of the evolution of plastid genomes within Ranunculaceae and across angiosperms. Thalictrum is unusual among the three sequenced Ranunculaceae plastid genomes in the loss of two genes infA and rpl32, which have been functionally transferred to the nucleus. In the case of rpl32 this represents the third documented independent transfer from the plastid to the nucleus with the other two transfers occurring in the unrelated angiosperm families Rhizophoraceae and Salicaceae. Furthermore, the transfer of rpl32 provides additional molecular evidence for the monophyly of the subfamily Thalictroideae.
Figures
Figure 1
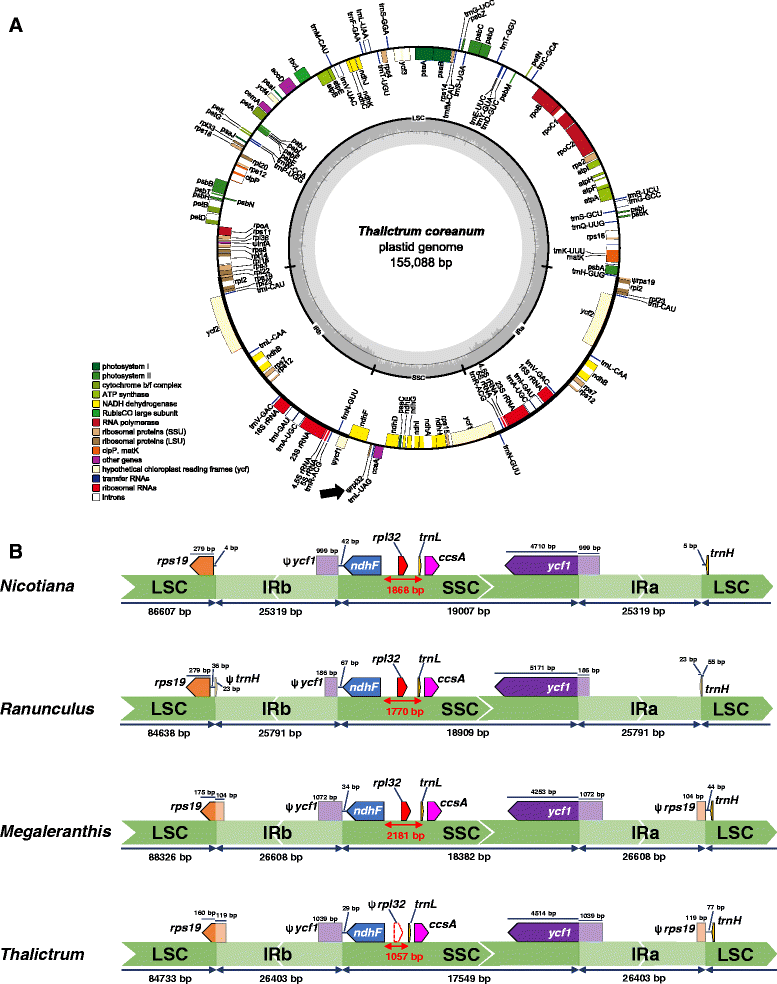
Circular gene map of Thalictrum coreanum plastome (A) and comparison of inverted repeat region of three plastomes from Ranunculaceae (B). A. Thick lines on inner circle indicate the inverted repeats (IRa and IRb, 26,403 bp), which separate the genome into small (SSC, 17,549 bp) and large (LSC, 84,733) single copy regions. Genes on the inside and outside of each map are transcribed clockwise and counterclockwise direction, respectively. The ring of bar graphs on the inner circle display GC content in dark grey. Ψ denotes a pseudogene and an arrow indicates the position of rpl32 pseudogene. B. Inverted repeat (IR) boundaries in three Ranunculaceae plastid genomes with Nicotiana tabacum as a reference genome are highlighted. Lengths of genes, large single copy (LSC), small single copy (SSC), and IRs are not to scale.
Figure 2
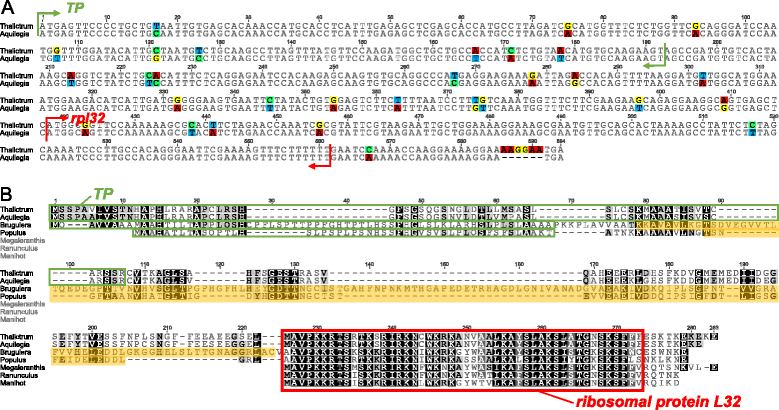
Alignment of the ribosomal protein L32 gene . A. Nucleotide sequence alignment of the nuclear-encoded rpl32 copies from Thalictrum and Aquilegia. B. Amino acid sequence alignment of the nuclear copies of rpl32 of Thalictrum, Aquilegia, and Populus with three plastid-encoded copies from related species. Green boxes indicate plastid transit peptides (TP) that were predicted using TargetP. Red box indicates a conserved domain of ribosomal protein L32. The shaded orange box indicates the putative Cu-Zn superoxide dismutase gene sequence.
Figure 3
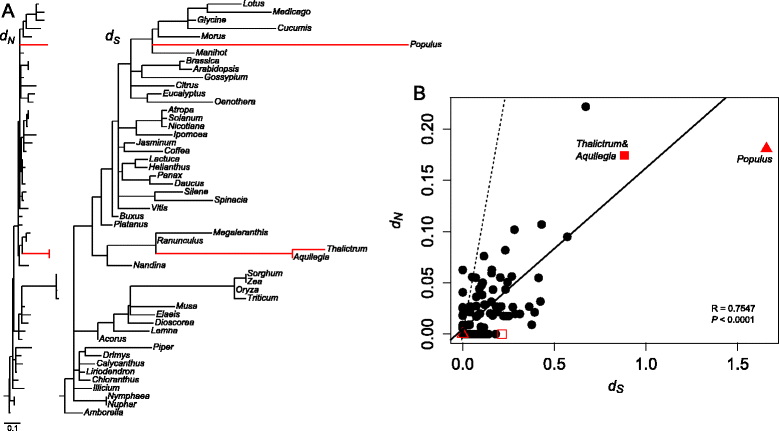
Nuclear- and plastid-encoded rpl32 divergence among selected angiosperms. A. Maximum likelihood trees showing nonsynonymous (d N) and synonymous (d S) substitution rates for plastid-encoded rpl32 genes with three nuclear-encoded copies. Red branches indicate the nuclear-encoded rpl32 copies. Trees are drawn to the same scale shown in the bottom left. B. Correlation of synonymous and nonsynonymous substitution rates of rpl32. Significance of fit was evaluated by a Pearson correlation coefficient in the R package. The solid line represents the regression, which was analyzed using d N and d S on all branches except for the Thalictrum (open square), Aquilegia (open triangle), Populus (triangle) terminal branches, and the branch leading to Thalictrum and Aquilegia (square). The dashed line indicates d N/d S ratio is equal to one.
Figure 4
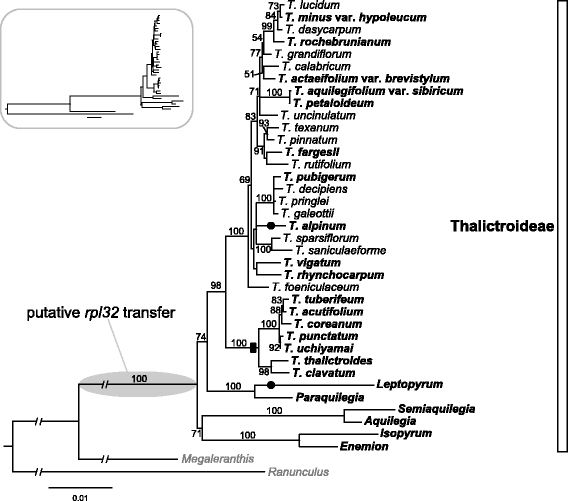
Phylogenetic relationships among 37 species of the subfamily Thalictroideae. Tree was constructed using nucleotide sequence of five plastid genes/regions (rbcL, ndhF, ndhA intron, trnL intron, and trnL-F intergenic spacer). The gray ellipse on node indicates putative transfer of rpl32 to the nucleus and black dots indicate the complete loss of rpl32 from plastid. Black rectangle on node indicates an indel event that is shared by members of the T. coreaum clade. Species in bold are those surveyed for loss of rpl32. Bootstrap support values > 50% are shown at nodes. Tree in box shows the original ML tree, which is broken (-//-) in the tree on right to make it easier to visualize. The circumscription of the subfamily Thalictroideae follows Wang et al. [23].
Figure 5
Nucleotide alignment of rpl32 gene/pseudogenes for Ranunculaceae. The top 15 sequences represent putative rpl32 pseudogenes for 15 Thalictrum species, the next five sequences are other genera within the subfamily Thalictroideae, and the bottom two sequences are representative species from outside of the subfamily Thalictroideae. Blue box shows an indel event that is shared by members of the T. coreaum clade.
Figure 6
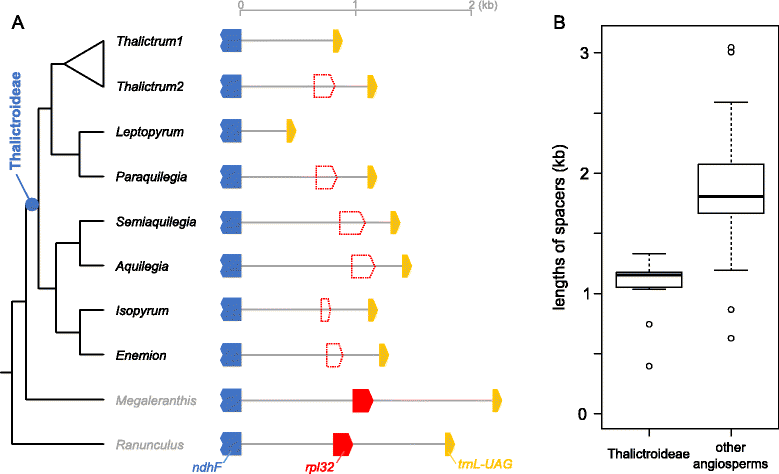
Length variation of intergenic spacer including rpl32 among species in the subfamily Thalictroideae. A. Schematic diagram of the regions surrounding the rpl32 gene in 22 sequenced species (right). In tree on left (reduced version of Figure 2), Thalictrum1 indicates Thalictrum alpinum and Thalictrum2 represents the remaining Thalictrum species. Dotted red boxes indicate the proportion of the remnant sequences from rpl32. B. Boxplot distribution of the lengths of the _ndhF_-trnL intergenic spacers between the subfamily Thalictroideae and other angiosperms that contain rpl32 gene (Additional file 2: Table S3).
Similar articles
- Complete plastid genome sequences of three Rosids (Castanea, Prunus, Theobroma): evidence for at least two independent transfers of rpl22 to the nucleus.
Jansen RK, Saski C, Lee SB, Hansen AK, Daniell H. Jansen RK, et al. Mol Biol Evol. 2011 Jan;28(1):835-47. doi: 10.1093/molbev/msq261. Epub 2010 Oct 8. Mol Biol Evol. 2011. PMID: 20935065 Free PMC article. - The evolutionary fate of rpl32 and rps16 losses in the Euphorbia schimperi (Euphorbiaceae) plastome.
Alqahtani AA, Jansen RK. Alqahtani AA, et al. Sci Rep. 2021 Apr 2;11(1):7466. doi: 10.1038/s41598-021-86820-z. Sci Rep. 2021. PMID: 33811236 Free PMC article. - Reconstructing evolution: gene transfer from plastids to the nucleus.
Bock R, Timmis JN. Bock R, et al. Bioessays. 2008 Jun;30(6):556-66. doi: 10.1002/bies.20761. Bioessays. 2008. PMID: 18478535 Review. - From chloroplasts to "cryptic" plastids: evolution of plastid genomes in parasitic plants.
Krause K. Krause K. Curr Genet. 2008 Sep;54(3):111-21. doi: 10.1007/s00294-008-0208-8. Epub 2008 Aug 12. Curr Genet. 2008. PMID: 18696071 Review.
Cited by
- The plastomes of Lepismium cruciforme (Vell.) Miq and Schlumbergera truncata (Haw.) Moran reveal tribe-specific rearrangements and the first loss of the trnT-GGU gene in Cactaceae.
Dalla Costa TP, Silva MC, de Santana Lopes A, Pacheco TG, da Silva GM, de Oliveira JD, de Baura VA, Balsanelli E, de Souza EM, de Oliveira Pedrosa F, Rogalski M. Dalla Costa TP, et al. Mol Biol Rep. 2024 Sep 4;51(1):957. doi: 10.1007/s11033-024-09871-1. Mol Biol Rep. 2024. PMID: 39230768 - The complete plastome of Thalictrum elegans Wall. ex Royle and its phylogenetic analysis.
Cheng R, Xu X, Chi X. Cheng R, et al. Mitochondrial DNA B Resour. 2024 Aug 20;9(8):1127-1131. doi: 10.1080/23802359.2024.2392756. eCollection 2024. Mitochondrial DNA B Resour. 2024. PMID: 39175481 Free PMC article. - Decoding the Chloroplast Genome of Tetrastigma (Vitaceae): Variations and Phylogenetic Selection Insights.
Zhu J, Huang Y, Chai W, Xia P. Zhu J, et al. Int J Mol Sci. 2024 Jul 29;25(15):8290. doi: 10.3390/ijms25158290. Int J Mol Sci. 2024. PMID: 39125860 Free PMC article. - Comparative and phylogenetic analyses of Loranthaceae plastomes provide insights into the evolutionary trajectories of plastome degradation in hemiparasitic plants.
Tang L, Wang T, Hou L, Zhang G, Deng M, Guo X, Ji Y. Tang L, et al. BMC Plant Biol. 2024 May 16;24(1):406. doi: 10.1186/s12870-024-05094-5. BMC Plant Biol. 2024. PMID: 38750463 Free PMC article.
References
- Bock R. Structure, function, and inheritance of plastid genomes. In: Bock R, editor. Cell and Molecular Biology of Plastids. Berlin Heidelberg: Springer; 2007. pp. 29–63.
- Jansen RK, Ruhlman TA. Plastid genomes of seed plants. In: Bock R, Knoop V, editors. Genomics of Chloroplasts and Mitochondria. New York: Springer; 2012. pp. 103–26.
- Glaser E, Soll J. Targeting signals and import machinery of plastids and plant mitochondria. In: Daniell H, Chase C, editors. Molecular Biology and Biotechnology of Plant Organelles: Chloroplasts and Mitochondria. New York: Springer; 2004. pp. 385–418.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases