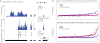Genomic variation. Impact of regulatory variation from RNA to protein - PubMed (original) (raw)
Genomic variation. Impact of regulatory variation from RNA to protein
Alexis Battle et al. Science. 2015.
Abstract
The phenotypic consequences of expression quantitative trait loci (eQTLs) are presumably due to their effects on protein expression levels. Yet the impact of genetic variation, including eQTLs, on protein levels remains poorly understood. To address this, we mapped genetic variants that are associated with eQTLs, ribosome occupancy (rQTLs), or protein abundance (pQTLs). We found that most QTLs are associated with transcript expression levels, with consequent effects on ribosome and protein levels. However, eQTLs tend to have significantly reduced effect sizes on protein levels, which suggests that their potential impact on downstream phenotypes is often attenuated or buffered. Additionally, we identified a class of cis QTLs that affect protein abundance with little or no effect on messenger RNA or ribosome levels, which suggests that they may arise from differences in posttranslational regulation.
Copyright © 2015, American Association for the Advancement of Science.
Figures
Figure 1. Comparisons of QTLs at three levels of gene regulation
a Many QTLs exhibit shared effects across mRNA, ribosome occupancy and protein. This example illustrates a shared QTL for the schlafen family member 5 (SLFN5) gene (24). The upper panels show mean sequence depth (per bp) for mRNA and ribosome occupancy, averaged among individuals with each genotype at the QTL SNP. The lower panel shows median log2 SILAC ratios at each detected peptide, relative to the shared internal standard. b. Replication rates between independently tested _cis_-QTLs for each phenotype pair, at FDR=10%. QTLs detected for the phenotype labeled on each row were tested in the phenotype listed for each column, considering only the 4,322 genes quantified in all three phenotypes. c. On average, eQTLs exhibit attenuated effects on protein abundance but not on ribosome occupancy. We used eQTLs detected by the GEUVADIS study to avoid ascertainment bias, and we polarized the alleles according to the direction of effect in GEUVADIS. The plot shows mean effect sizes and standard errors on the means, measured as expected fold-change per allele copy on a log2 scale.
Figure 2. Protein-specific and RNA-specific QTLs
a An example of a protein-specific QTL, for the apolipoprotein L, 2 (APOL2) gene, detected by both the interaction model and the conditional models, indicating both larger effect (LRT, P = 3.3×10-6, interaction model; P = 5.1×10-13, conditional model) in protein than mRNA, and that the effect on protein is not mediated by either mRNA or ribosome occupancy (LRT, P = 2.1×10-12, conditional model). Plotting details as in Figure 1A. While the causal variant underlying this pQTL is unknown, several linked variants near the 3′ end of APOL2 are all strongly associated with protein levels, including rs8142325 shown here and missense variant rs7285167 (βg = 0.83, P = 9.8×10-9; LRT, P = 2.1×10-5, interaction model; P = 5.5×10-13, conditional model). b. Effect sizes for ribosome occupancy tend to track with RNA, not protein. Top panel: effect sizes in all three phenotypes are shown for protein-specific QTLs. Effect sizes were estimated using linear regression in each of the phenotypes independently. The signs of the effects were set to be positive in protein. Solid lines reflect predicted effects based on a linear model. Bottom panel: Similarly, effect sizes in all three phenotypes for esQTLs. Here, signs of the effects were set to be positive in RNA.
Comment in
- Gene expression. Statistics requantitates the central dogma.
Li JJ, Biggin MD. Li JJ, et al. Science. 2015 Mar 6;347(6226):1066-7. doi: 10.1126/science.aaa8332. Science. 2015. PMID: 25745146 No abstract available.
Similar articles
- The impact of common variants on gene expression in the human brain: from RNA to protein to schizophrenia risk.
Liang Q, Jiang Y, Shieh AW, Zhou D, Chen R, Wang F, Xu M, Niu M, Wang X, Pinto D, Wang Y, Cheng L, Vadukapuram R, Zhang C, Grennan K, Giase G; PsychENCODE Consortium; White KP, Peng J, Li B, Liu C, Chen C, Wang SH. Liang Q, et al. bioRxiv [Preprint]. 2023 Nov 10:2023.06.04.543603. doi: 10.1101/2023.06.04.543603. bioRxiv. 2023. PMID: 37873195 Free PMC article. Preprint. - The abundance of cis-acting loci leading to differential allele expression in F1 mice and their relationship to loci harboring genes affecting complex traits.
Yeo S, Hodgkinson CA, Zhou Z, Jung J, Leung M, Yuan Q, Goldman D. Yeo S, et al. BMC Genomics. 2016 Aug 11;17(1):620. doi: 10.1186/s12864-016-2922-9. BMC Genomics. 2016. PMID: 27515598 Free PMC article. - Statistically and functionally fine-mapped blood eQTLs and pQTLs from 1,405 humans reveal distinct regulation patterns and disease relevance.
Wang QS, Hasegawa T, Namkoong H, Saiki R, Edahiro R, Sonehara K, Tanaka H, Azekawa S, Chubachi S, Takahashi Y, Sakaue S, Namba S, Yamamoto K, Shiraishi Y, Chiba K, Tanaka H, Makishima H, Nannya Y, Zhang Z, Tsujikawa R, Koike R, Takano T, Ishii M, Kimura A, Inoue F, Kanai T, Fukunaga K, Ogawa S, Imoto S, Miyano S, Okada Y; Japan COVID-19 Task Force. Wang QS, et al. Nat Genet. 2024 Oct;56(10):2054-2067. doi: 10.1038/s41588-024-01896-3. Epub 2024 Sep 24. Nat Genet. 2024. PMID: 39317738 Free PMC article. - The study of eQTL variations by RNA-seq: from SNPs to phenotypes.
Majewski J, Pastinen T. Majewski J, et al. Trends Genet. 2011 Feb;27(2):72-9. doi: 10.1016/j.tig.2010.10.006. Epub 2010 Nov 29. Trends Genet. 2011. PMID: 21122937 Review. - QTL Analysis Beyond eQTLs.
Wen J, Nodzak C, Shi X. Wen J, et al. Methods Mol Biol. 2020;2082:201-210. doi: 10.1007/978-1-0716-0026-9_14. Methods Mol Biol. 2020. PMID: 31849017 Review.
Cited by
- A pooling-based approach to mapping genetic variants associated with DNA methylation.
Kaplow IM, MacIsaac JL, Mah SM, McEwen LM, Kobor MS, Fraser HB. Kaplow IM, et al. Genome Res. 2015 Jun;25(6):907-17. doi: 10.1101/gr.183749.114. Epub 2015 Apr 24. Genome Res. 2015. PMID: 25910490 Free PMC article. - Long non-coding RNAs in brain tumors: roles and potential as therapeutic targets.
Kim SH, Lim KH, Yang S, Joo JY. Kim SH, et al. J Hematol Oncol. 2021 May 12;14(1):77. doi: 10.1186/s13045-021-01088-0. J Hematol Oncol. 2021. PMID: 33980320 Free PMC article. Review. - Regulatory Nucleotide Sequence Signals for Expression of the Genes Encoding Ribosomal Proteins.
Ryu J, Lee C. Ryu J, et al. Front Genet. 2020 Jun 5;11:501. doi: 10.3389/fgene.2020.00501. eCollection 2020. Front Genet. 2020. PMID: 32655613 Free PMC article. - The role of regulatory variation in complex traits and disease.
Albert FW, Kruglyak L. Albert FW, et al. Nat Rev Genet. 2015 Apr;16(4):197-212. doi: 10.1038/nrg3891. Epub 2015 Feb 24. Nat Rev Genet. 2015. PMID: 25707927 Review. - Striatal cell-type-specific molecular signatures reveal therapeutic targets in a model of dystonia.
Roman KM, Dinasarapu AR, Cherian S, Fan X, Donsante Y, Aravind N, Chan CS, Jinnah HA, Hess EJ. Roman KM, et al. bioRxiv [Preprint]. 2024 Oct 7:2024.10.07.617010. doi: 10.1101/2024.10.07.617010. bioRxiv. 2024. PMID: 39415987 Free PMC article. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
- F32HG006972/HG/NHGRI NIH HHS/United States
- R01 MH084703/MH/NIMH NIH HHS/United States
- F32 HG006972/HG/NHGRI NIH HHS/United States
- GM077959/GM/NIGMS NIH HHS/United States
- R01 GM077959/GM/NIGMS NIH HHS/United States
- HG007036/HG/NHGRI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U01 HG007036/HG/NHGRI NIH HHS/United States
- MH084703/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases