DNA double strand break repair pathway choice: a chromatin based decision? - PubMed (original) (raw)
DNA double strand break repair pathway choice: a chromatin based decision?
T Clouaire et al. Nucleus. 2015.
Abstract
DNA double-strand breaks (DSBs) are highly toxic lesions that can be rapidly repaired by 2 main pathways, namely Homologous Recombination (HR) and Non Homologous End Joining (NHEJ). The choice between these pathways is a critical, yet not completely understood, aspect of DSB repair. We recently found that distinct DSBs induced across the genome are not repaired by the same pathway. Indeed, DSBs induced in active genes, naturally enriched in the trimethyl form of histone H3 lysine 36 (H3K36me3), are channeled to repair by HR, in a manner depending on SETD2, the major H3K36 trimethyltransferase. Here, we propose that these findings may be generalized to other types of histone modifications and repair machineries thus defining a "DSB repair choice histone code". This "decision making" function of preexisting chromatin structure in DSB repair could connect the repair pathway used to the type and function of the damaged region, not only contributing to genome stability but also to its diversity.
Keywords: DNA double strand breaks; DSB pathway choice; chromatin, trimethyl H3K36; homologous recombination.
Figures
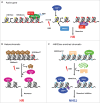
Figure 1.
Preexistent chromatin structure influences DSB repair. (A) Actively transcribed genes are enriched for the transcription elongation mark H3K36me3 (red circles) by the action of SETD2 associating with elongating RNA Pol II. Histone acetylation on H4K16 (H4K16ac, yellow circles) is also enriched on active transcription units. Upon DSB induction, LEDGF-mediated CtIP recruitment, via H3K36me3 recognition, will favor resection and channel repair toward HR. H4K16ac will impede 53BP1 accumulation near DSBs, which will favor resection and BRCA1 dependent repair. (B) Condensed heterochromatin is characterized by H3K9me3 enrichment (gray trapezoids), the histone mark bound by HP1 via its chromodomain. Following DSBs, HP1 is evicted, allowing for Tip60 to interact with H3K9me3 through its own chromodomain. Tip60-dependant acetylation will favor chromatin remodeling and nucleosome removal, and probably resection, required for HR repair of DSB occurring in heterochromatin. (C) H4K20 mono and dimethylation (blue circles) are abundant modifications in mammalian genomes. Upon DSB, H4K20me1/2 is unmasked, via L3MBTL1 eviction or KDM4A degradation, DSB induced structural changes or histone modifications such as H2AK15ub (purple squares). This favors 53BP1 recruitment near the break sites, inhibits resection, allowing DSB repair by NHEJ.
Figure 2.
Potential mechanisms for preexistent chromatin structure in addressing repair pathways. (A) Following DSB and initial recognition by end binding proteins, such as Ku or the MRN complex, chromatin modifications already present at the break will stabilize component of a specific DNA repair pathway and/or inhibit association of component from another pathway. (B) DSB induces unmasking of existing histone marks by chromatin structure remodeling and/or chromatin reader eviction, making these modifications available for recognition by DNA repair factors. (C) In undamaged chromatin, repair proteins may preferentially interact with certain region of the genome based on their specific chromatin signature, facilitating their loading upon DSB induction.
Similar articles
- Di- and tri-methylation of histone H3K36 play distinct roles in DNA double-strand break repair.
Chen R, Zhao MJ, Li YM, Liu AH, Wang RX, Mei YC, Chen X, Du HN. Chen R, et al. Sci China Life Sci. 2024 Jun;67(6):1089-1105. doi: 10.1007/s11427-024-2543-9. Epub 2024 Feb 29. Sci China Life Sci. 2024. PMID: 38842635 - The Chromatin Landscape around DNA Double-Strand Breaks in Yeast and Its Influence on DNA Repair Pathway Choice.
Frigerio C, Di Nisio E, Galli M, Colombo CV, Negri R, Clerici M. Frigerio C, et al. Int J Mol Sci. 2023 Feb 7;24(4):3248. doi: 10.3390/ijms24043248. Int J Mol Sci. 2023. PMID: 36834658 Free PMC article. Review. - Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks.
Aymard F, Bugler B, Schmidt CK, Guillou E, Caron P, Briois S, Iacovoni JS, Daburon V, Miller KM, Jackson SP, Legube G. Aymard F, et al. Nat Struct Mol Biol. 2014 Apr;21(4):366-74. doi: 10.1038/nsmb.2796. Epub 2014 Mar 23. Nat Struct Mol Biol. 2014. PMID: 24658350 Free PMC article. - Timely double-strand break repair and pathway choice in pericentromeric heterochromatin depend on the histone demethylase dKDM4A.
Janssen A, Colmenares SU, Lee T, Karpen GH. Janssen A, et al. Genes Dev. 2019 Jan 1;33(1-2):103-115. doi: 10.1101/gad.317537.118. Epub 2018 Dec 21. Genes Dev. 2019. PMID: 30578303 Free PMC article. - Regulation of DNA double-strand break repair pathway choice.
Shrivastav M, De Haro LP, Nickoloff JA. Shrivastav M, et al. Cell Res. 2008 Jan;18(1):134-47. doi: 10.1038/cr.2007.111. Cell Res. 2008. PMID: 18157161 Review.
Cited by
- Chromatin context-dependent effects of epigenetic drugs on CRISPR-Cas9 editing.
Schep R, Trauernicht M, Vergara X, Friskes A, Morris B, Gregoricchio S, Manzo SG, Zwart W, Beijersbergen RL, Medema RH, van Steensel B. Schep R, et al. Nucleic Acids Res. 2024 Aug 27;52(15):8815-8832. doi: 10.1093/nar/gkae570. Nucleic Acids Res. 2024. PMID: 38953163 Free PMC article. - Phasor Histone FLIM-FRET Microscopy Maps Nuclear-Wide Nanoscale Chromatin Architecture With Respect to Genetically Induced DNA Double-Strand Breaks.
Lou J, Solano A, Liang Z, Hinde E. Lou J, et al. Front Genet. 2021 Dec 10;12:770081. doi: 10.3389/fgene.2021.770081. eCollection 2021. Front Genet. 2021. PMID: 34956323 Free PMC article. - H3K36me3, message from chromatin to DNA damage repair.
Sun Z, Zhang Y, Jia J, Fang Y, Tang Y, Wu H, Fang D. Sun Z, et al. Cell Biosci. 2020 Jan 31;10:9. doi: 10.1186/s13578-020-0374-z. eCollection 2020. Cell Biosci. 2020. PMID: 32021684 Free PMC article. Review. - Complex interactions between the DNA-damage response and mammalian telomeres.
Arnoult N, Karlseder J. Arnoult N, et al. Nat Struct Mol Biol. 2015 Nov;22(11):859-66. doi: 10.1038/nsmb.3092. Nat Struct Mol Biol. 2015. PMID: 26581520 Free PMC article. Review. - Non-redundant Functions of ATM and DNA-PKcs in Response to DNA Double-Strand Breaks.
Caron P, Choudjaye J, Clouaire T, Bugler B, Daburon V, Aguirrebengoa M, Mangeat T, Iacovoni JS, Álvarez-Quilón A, Cortés-Ledesma F, Legube G. Caron P, et al. Cell Rep. 2015 Nov 24;13(8):1598-609. doi: 10.1016/j.celrep.2015.10.024. Epub 2015 Nov 12. Cell Rep. 2015. PMID: 26586426 Free PMC article.
References
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell 2010; 40:179-204; PMID:20965415; http://dx.doi.org/10.1016/j.molcel.2010.09.019 - DOI - PMC - PubMed
- Betermier M, Bertrand P, Lopez BS. Is non-homologous end-joining really an inherently error-prone process? PLoS Genetics 2014; 10:e1004086; PMID:24453986; http://dx.doi.org/10.1371/journal.pgen.1004086 - DOI - PMC - PubMed
- Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol 2010; 17:410-6; PMID:20208544; http://dx.doi.org/10.1038/nsmb.1773 - DOI - PMC - PubMed
- Ghezraoui H, Piganeau M, Renouf B, Renaud JB, Sallmyr A, Ruis B, Oh S, Tomkinson AE, Hendrickson EA, Giovannangeli C, et al. . Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol Cell 2014; 55:829-42; PMID:25201414; http://dx.doi.org/10.1016/j.molcel.2014.08.002 - DOI - PMC - PubMed
- Rai R, Zheng H, He H, Luo Y, Multani A, Carpenter PB, Chang S. The function of classical and alternative non-homologous end-joining pathways in the fusion of dysfunctional telomeres. EMBO J 2010; 29:2598-610; PMID:20588252; http://dx.doi.org/10.1038/emboj.2010.142 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources