Spatial enhancer clustering and regulation of enhancer-proximal genes by cohesin - PubMed (original) (raw)
Spatial enhancer clustering and regulation of enhancer-proximal genes by cohesin
Elizabeth Ing-Simmons et al. Genome Res. 2015 Apr.
Abstract
In addition to mediating sister chromatid cohesion during the cell cycle, the cohesin complex associates with CTCF and with active gene regulatory elements to form long-range interactions between its binding sites. Genome-wide chromosome conformation capture had shown that cohesin's main role in interphase genome organization is in mediating interactions within architectural chromosome compartments, rather than specifying compartments per se. However, it remains unclear how cohesin-mediated interactions contribute to the regulation of gene expression. We have found that the binding of CTCF and cohesin is highly enriched at enhancers and in particular at enhancer arrays or "super-enhancers" in mouse thymocytes. Using local and global chromosome conformation capture, we demonstrate that enhancer elements associate not just in linear sequence, but also in 3D, and that spatial enhancer clustering is facilitated by cohesin. The conditional deletion of cohesin from noncycling thymocytes preserved enhancer position, H3K27ac, H4K4me1, and enhancer transcription, but weakened interactions between enhancers. Interestingly, ∼ 50% of deregulated genes reside in the vicinity of enhancer elements, suggesting that cohesin regulates gene expression through spatial clustering of enhancer elements. We propose a model for cohesin-dependent gene regulation in which spatial clustering of enhancer elements acts as a unified mechanism for both enhancer-promoter "connections" and "insulation."
© 2015 Ing-Simmons et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
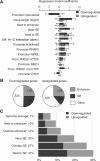
Figure 1.
Cohesin facilitates the regulated expression of genes located near enhancer elements. (A) Enhancers and super-enhancers (Supplemental Fig. S1) emerge as major determinants of cohesin-dependent gene regulation. The “Next to enhancer” category includes genes that are nearest neighbors of conventional enhancers. The “Near SE” category includes genes that are positioned within 40 kb of an SE, including genes that overlap SEs and nearest neighbors of SEs. “Next to SE” includes only nearest neighbors of SEs. “Diff. Hi-C interaction” indicates genes in 100-kb regions that interact differentially in control and _Rad21_-deficient thymocytes (Seitan et al. 2013). Other categories refer to promoter binding of the listed factors (TSS ± 2.5 kb). Multivariate multinomial regression analysis was done as described in Seitan et al. (2013). The statistical significance of regression model coefficients is indicated and only significant associations are shown. (B) Proximity to enhancers accounts for ∼50% of genes deregulated in _Rad21_-deficient thymocytes. (Enhancer) Genes that are nearest neighbors or overlap conventional enhancers; (SE) genes ±40 kb of a super-enhancer, overlapping a super-enhancer, or that are nearest neighbors of super-enhancers. (C) Cohesin is required for the regulation of genes near enhancers and super-enhancers. (Genome average) Expressed genes (17,003 genes); (Next to enhancer) nearest neighbors of conventional enhancers (3540 genes); (Overlapping enhancer) genes that overlap conventional enhancers (198 genes); (Near SE) ±40 kb of a super-enhancer (1036 genes); (Overlapping SE) overlap a super-enhancer (493 genes); (Next to SE) nearest neighbors of super-enhancers (447 genes). Additional data are shown in Supplemental Table 1.
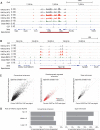
Figure 2.
Enhancer elements are maintained in cohesin-deficient thymocytes. The Cd8 (A) and the Ppp1r16b region (B) illustrate the maintenance of H3K27ac in cohesin-deficient thymocytes. Binding of the cohesin subunits RAD21 and SMC1A as well as CTCF is shown for reference (see below). (C) Rad21 deletion does not abolish H3K27ac marking of conventional enhancers (left), newly established developmentally regulated enhancers (red, middle), or super-enhancers (right). (D) Enhancers and super-enhancers were stratified according to the ratio of H3K27ac in _Rad21_-deficient thymocytes over wild type. The frequency of deregulated genes is shown for the top, middle, and lower third of enhancers (left) and super-enhancers (right).
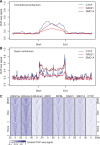
Figure 3.
Enhancer elements are enriched for CTCF and cohesin binding. (A) CTCF, RAD21, and SMC1A ChIP-seq signal enrichment in thymocytes at conventional enhancers. Enhancers were defined based on H2K27ac ChIP-seq data (Methods) and enhancer length was normalized in order to align the start and end of the enhancer. (B) CTCF, RAD21, and SMC1A ChIP-seq signal enrichment in thymocytes at super-enhancers. Super-enhancers were defined based on H2K27ac ChIP-seq data (Methods), enhancer length was normalized in order to align the start and end of the super-enhancers, and flanking regions of equal size to the super-enhancer are shown for reference. “Start” and “end” are based on the genomic coordinates. (C) Classification of super-enhancers on the basis of CTCF binding at both boundaries, one boundary, or neither boundary. Heatmaps of ChIP-seq signal enrichment in 100-kb windows around super-enhancer centers, grouped according to CTCF binding at the super-enhancer boundaries. Histone modifications, the cohesin loading factor NIPBL, and the cohesin subunits RAD21 and SMC1A are shown for reference.
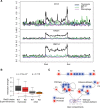
Figure 4.
Cohesin mediates the spatial clustering of enhancer elements. (A) The mouse Cd3 super-enhancer is flanked and punctuated by CTCF and cohesin binding (refer to Fig. 2A,B for additional examples of the relationship between super-enhancers, CTCF, and cohesin). Restriction fragments used for 3C analysis are indicated by gray bars. “A” is a proximity ligation control that demonstrates comparable efficiency of 3C experiments. Enhancer-enhancer interactions B, C, and D across the Cd3 super-enhancer were significantly reduced in _Rad21-_deficient (red) compared to control (black) thymocytes. n = 3, mean ± SD, (*) P < 0.05. (B) Long-range interactions between the Cd3 super-enhancer and enhancer elements outside the Cd3 locus. “E” is a control used to demonstrate background interactions with a downstream genomic fragment lacking H3K27ac marks and CTCF binding. The position of the Cd3 super-enhancer is marked by dashed lines. Interactions F, G, H, I, and J link the Cd3 super-enhancer with downstream enhancer elements outside the Cd3 region and were significantly reduced in _Rad21-_deficient (red) compared to control (black) thymocytes. n = 3, mean ± SD, (*) P < 0.05. (C) Structured interaction matrix analysis (SIMA) of long-range interactions between chromatin features based on Hi-C data for control (gray) and _Rad21_-deficient (red) thymocytes. Interactions between constituent elements of super-enhancers (left) were analyzed by SIMA within super-enhancer regions sized 100 kb and larger. Interactions between other chromatin features were analyzed by SIMA within open chromatin compartments. Note that chromatin features show increased self-interactions, while interactions of random regions (Seitan et al. 2013) conformed to the level of interactions predicted by a background model based on genomic distance and sequencing depth (dashed red line). “Interaction strength” refers to the strength of interactions between 10-kb regions within super-enhancers (SEs) or 10-kb regions within chromosomal compartments (Enh, RAD21 sites, TSS, and random regions after normalization to the background model). _P_-values shown are from a Wilcoxon signed-rank test. (D) Reduced enhancer-enhancer interactions in _Rad21_-deficient thymocytes based on SIMA analysis of Hi-C data are compared for all open chromosomal compartments and separately for compartments with or without super-enhancers. _P_-values shown are from a Wilcoxon signed-rank test.
Figure 5.
Cell-type specificity of CTCF association and spatial clustering of super-enhancers. (A) CTCF (top), RAD21 (middle), and SMC1A (bottom) ChIP-seq signal in thymocytes at super-enhancers that are active in thymocytes (black), macrophages (purple), myoblasts (blue), or ES cells (green). (B) Preferential interactions within cell-type-specific super-enhancers. SIMA analysis of thymocyte Hi-C data was used to compare interactions within super-enhancers active in thymocytes or other cell types. (Left) Hi-C interactions in control (gray) and _Rad21_-deficient thymocytes (red) within thymocyte super-enhancers sized 50 kb or more (n = 105). (Right) Hi-C interactions in WT (light gray) and _Rad21_-deficient thymocytes (orange) within super-enhancers of 50 kb or more that are active in pro-B cells, macrophages, or ES cells (n = 20) (Whyte et al. 2013). _P_-values shown are from a Wilcoxon signed-rank test. (C) Model for the impact of cohesin-dependent enhancer-enhancer interactions on gene expression. Spatial clustering between enhancer elements can affect promoter activity positively and negatively. The promoters P1 and P3 are both distal to a super-enhancer. P1 is isolated as a result of the spatial clustering between the enhancer elements within the SE, while P3 is connected to the super-enhancer by enhancer-enhancer contacts (left). Removal of cohesin (right) decreases the spatial constraint on enhancer elements so that P1 is contacted more readily by enhancer elements, while P3 dissociates from the super-enhancer.
Similar articles
- Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments.
Seitan VC, Faure AJ, Zhan Y, McCord RP, Lajoie BR, Ing-Simmons E, Lenhard B, Giorgetti L, Heard E, Fisher AG, Flicek P, Dekker J, Merkenschlager M. Seitan VC, et al. Genome Res. 2013 Dec;23(12):2066-77. doi: 10.1101/gr.161620.113. Epub 2013 Sep 3. Genome Res. 2013. PMID: 24002784 Free PMC article. - Cohesin mediates transcriptional insulation by CCCTC-binding factor.
Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM. Wendt KS, et al. Nature. 2008 Feb 14;451(7180):796-801. doi: 10.1038/nature06634. Epub 2008 Jan 30. Nature. 2008. PMID: 18235444 - Long-Range Chromosome Interactions Mediated by Cohesin Shape Circadian Gene Expression.
Xu Y, Guo W, Li P, Zhang Y, Zhao M, Fan Z, Zhao Z, Yan J. Xu Y, et al. PLoS Genet. 2016 May 2;12(5):e1005992. doi: 10.1371/journal.pgen.1005992. eCollection 2016 May. PLoS Genet. 2016. PMID: 27135601 Free PMC article. - Cohesin and CTCF: cooperating to control chromosome conformation?
Gause M, Schaaf CA, Dorsett D. Gause M, et al. Bioessays. 2008 Aug;30(8):715-8. doi: 10.1002/bies.20787. Bioessays. 2008. PMID: 18623068 Review. - Insulators and domains of gene expression.
Ali T, Renkawitz R, Bartkuhn M. Ali T, et al. Curr Opin Genet Dev. 2016 Apr;37:17-26. doi: 10.1016/j.gde.2015.11.009. Epub 2016 Jan 20. Curr Opin Genet Dev. 2016. PMID: 26802288 Review.
Cited by
- Single-Nucleus RNA Sequencing Reveals Loss of Distal Convoluted Tubule 1 Renal Tubules in HIV Viral Protein R Transgenic Mice.
Latt KZ, Yoshida T, Shrivastav S, Abedini A, Reece JM, Sun Z, Lee H, Okamoto K, Dagur P, Ishimoto Y, Heymann J, Zhao Y, Chung JY, Hewitt S, Jose PA, Lee K, He JC, Winkler CA, Knepper MA, Kino T, Rosenberg AZ, Susztak K, Kopp JB. Latt KZ, et al. Am J Pathol. 2024 Oct;194(10):1844-1856. doi: 10.1016/j.ajpath.2024.06.006. Epub 2024 Jul 18. Am J Pathol. 2024. PMID: 39032602 - Functional genomic approaches to elucidate the role of enhancers during development.
Ryan GE, Farley EK. Ryan GE, et al. Wiley Interdiscip Rev Syst Biol Med. 2020 Mar;12(2):e1467. doi: 10.1002/wsbm.1467. Epub 2019 Dec 5. Wiley Interdiscip Rev Syst Biol Med. 2020. PMID: 31808313 Free PMC article. Review. - Live-cell three-dimensional single-molecule tracking reveals modulation of enhancer dynamics by NuRD.
Basu S, Shukron O, Hall D, Parutto P, Ponjavic A, Shah D, Boucher W, Lando D, Zhang W, Reynolds N, Sober LH, Jartseva A, Ragheb R, Ma X, Cramard J, Floyd R, Balmer J, Drury TA, Carr AR, Needham LM, Aubert A, Communie G, Gor K, Steindel M, Morey L, Blanco E, Bartke T, Di Croce L, Berger I, Schaffitzel C, Lee SF, Stevens TJ, Klenerman D, Hendrich BD, Holcman D, Laue ED. Basu S, et al. Nat Struct Mol Biol. 2023 Nov;30(11):1628-1639. doi: 10.1038/s41594-023-01095-4. Epub 2023 Sep 28. Nat Struct Mol Biol. 2023. PMID: 37770717 Free PMC article. - Multiplexed capture of spatial configuration and temporal dynamics of locus-specific 3D chromatin by biotinylated dCas9.
Liu X, Chen Y, Zhang Y, Liu Y, Liu N, Botten GA, Cao H, Orkin SH, Zhang MQ, Xu J. Liu X, et al. Genome Biol. 2020 Mar 5;21(1):59. doi: 10.1186/s13059-020-01973-w. Genome Biol. 2020. PMID: 32138752 Free PMC article. - Hotspots of aberrant enhancer activity punctuate the colorectal cancer epigenome.
Cohen AJ, Saiakhova A, Corradin O, Luppino JM, Lovrenert K, Bartels CF, Morrow JJ, Mack SC, Dhillon G, Beard L, Myeroff L, Kalady MF, Willis J, Bradner JE, Keri RA, Berger NA, Pruett-Miller SM, Markowitz SD, Scacheri PC. Cohen AJ, et al. Nat Commun. 2017 Feb 7;8:14400. doi: 10.1038/ncomms14400. Nat Commun. 2017. PMID: 28169291 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- Wellcome Trust/United Kingdom
- MC_U120027516/MRC_/Medical Research Council/United Kingdom
- MC_UP_1102/1/MRC_/Medical Research Council/United Kingdom
- R01 HG003143/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases