Innate immune restriction and antagonism of viral RNA lacking 2׳-O methylation - PubMed (original) (raw)
Review
Innate immune restriction and antagonism of viral RNA lacking 2׳-O methylation
Jennifer L Hyde et al. Virology. 2015 May.
Abstract
N-7 and 2'-O methylation of host cell mRNA occurs in the nucleus and results in the generation of cap structures (cap 0, m(7)GpppN; cap 1, m(7)GpppNm) that control gene expression by modulating nuclear export, splicing, turnover, and protein synthesis. Remarkably, RNA cap modification also contributes to mammalian cell host defense as viral RNA lacking 2'-O methylation is sensed and inhibited by IFIT1, an interferon (IFN) stimulated gene (ISG). Accordingly, pathogenic viruses that replicate in the cytoplasm have evolved mechanisms to circumvent IFIT1 restriction and facilitate infection of mammalian cells. These include: (a) generating cap 1 structures on their RNA through cap-snatching or virally-encoded 2'-O methyltransferases, (b) using cap-independent means of translation, or (c) using RNA secondary structural motifs to antagonize IFIT1 binding. This review will discuss new insights as to how specific modifications at the 5'-end of viral RNA modulate host pathogen recognition responses to promote infection and disease.
Keywords: Immune evasion; Innate immunity; Interferon; Methylation; RNA structure; Viral pathogenesis.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
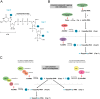
Fig. 1
Overview of cap 1 structure formation in eukaryotic and viral systems. (A) Chemical structure of the G-cap and methylation sites which form cap 0, cap 1, and cap 2 structures. (B) The eukaryotic canonical capping pathway. Newly transcribed RNA (pppNp-RNA) is cleaved by RNA triphosphatase at the γ-phosphate to yield diphosphate RNA (ppNp-RNA). Guanylyltransferase, which forms part of the bifunctional capping enzyme (hCAP) that also encodes triphosphatase activity, then hydrolyzes the α-phosphate from GTP and transfers the GMP moiety to the ppNp-RNA acceptor to generate GpppNp. GpppNp-RNA is methylated by the N-7 MTase, which transfers a methyl group from an S-adenosyl-
l
-methionine (AdoMet) donor to the unmethylated G-cap acceptor, producing cap 0 RNA (m7GpppNp) and S-adenosyl-
l
-homocysteine (AdoHcy) as a by-product. The _2′-O_-ribose MTase (hMTr1) then transfers a methyl group from the AdoMet donor to the first and second nucleotides of cap 0 RNA to generate cap 1 and cap 2 structures, respectively. (C) Generation of viral cap 1 structures through non-canonical capping and methylation pathways. Analogous to eukaryotic RNA methylation, some viruses (e.g. coronavirus and poxvirus) catalyze cap 0 and cap 1 formation separately via two distinct enzymes or enzymes complexes (left). In contrast, other viruses (e.g. flavivirus and reovirus) catalyze N-7 and 2′-O methylation sequentially via a single enzyme (right). Although the cap 1 end product is identical between host and viral transcripts structural differences in the viral and eukaryotic enzymes that catalyze these reactions and their distinct modes of ligand binding make viral 2′-O MTases potential targets for inhibitor design.
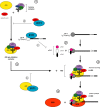
Fig. 2
Ifit1 inhibits diverse aspects of RNA translation. Canonical cap-dependent translation proceeds as follows: (1) the 40S ribosomal subunit complexes with eIF3 and the ternary complex (eIF2-GTP-Met-tRNA) to form the 43S pre-initiation complex. (2) eIF4E associates with the cap and recruits the eIF4F cap-binding complex (eIF4A, eIF4E, eIF4G). (3) eIF4F facilitates unwinding of the RNA and the 43S pre-initiation complex is recruited to form the 48S complex. (4) The 48S complex scans along the mRNA for the AUG initiator codon. (5) The 60S ribosomal subunit is recruited to the 48S complex to form the 80S initiator complex, which translates the mRNA. Ifit1/IFIT1 binds to subunits of the eIF3 complex (a) to prevent ternary complex formation or 43S formation and recruitment (Guo et al., 2000a, Hui et al., 2003, Hui et al., 2005, Kumar et al., 2014, Wang et al., 2003). (b) IFIT1 interaction with the 40S ribosomal subunit abrogates 48S formation (Kumar et al., 2014). (c) Ifit1/IFIT1 directly associates with cap 0 viral RNA and inhibits recruitment of eIF4E and eIF4F to RNA.
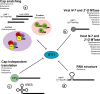
Fig. 3
Viral mechanisms of Ifit1 evasion and antagonism. Viruses generate cap 1 structures through (a) cap-snatching (orthomyxoviurses, bunyaviruses, arenaviruses) whereby cap 1 is cleaved from host mRNA transcripts and used as a primer for viral RNA transcription. In contrast to orthomyxoviruses, which cap-snatch from mRNA in the nucleus, bunyaviruses associate with and sequester cap 1 containing nonsense-mRNA transcripts in cellular P bodies where N protein protects cap 1 from degradation. Some viruses mimic cap 1 structures via virally-encoded 2′-O MTases (flaviviruses, coronaviruses, rhabdoviruses, paramyxoviruses, reoviruses, and poxviruses). Other viruses (picornaviruses, caliciviruses, and hepaciviruses) circumvent Ifit1-mediated restriction by using non-canonical cap-like structures (VPg) at their 5′ end or IRES-mediated cap-independent translation (b). Alphaviruses antagonize Ifit1 function directly by inhibiting association with viral RNA through the generation of stable secondary structures in the 5′-UTR (c).
Similar articles
- Structure of human IFIT1 with capped RNA reveals adaptable mRNA binding and mechanisms for sensing N1 and N2 ribose 2'-O methylations.
Abbas YM, Laudenbach BT, Martínez-Montero S, Cencic R, Habjan M, Pichlmair A, Damha MJ, Pelletier J, Nagar B. Abbas YM, et al. Proc Natl Acad Sci U S A. 2017 Mar 14;114(11):E2106-E2115. doi: 10.1073/pnas.1612444114. Epub 2017 Mar 1. Proc Natl Acad Sci U S A. 2017. PMID: 28251928 Free PMC article. - A viral RNA structural element alters host recognition of nonself RNA.
Hyde JL, Gardner CL, Kimura T, White JP, Liu G, Trobaugh DW, Huang C, Tonelli M, Paessler S, Takeda K, Klimstra WB, Amarasinghe GK, Diamond MS. Hyde JL, et al. Science. 2014 Feb 14;343(6172):783-7. doi: 10.1126/science.1248465. Epub 2014 Jan 30. Science. 2014. PMID: 24482115 Free PMC article. - 2'-O methylation of the viral mRNA cap by West Nile virus evades ifit1-dependent and -independent mechanisms of host restriction in vivo.
Szretter KJ, Daniels BP, Cho H, Gainey MD, Yokoyama WM, Gale M Jr, Virgin HW, Klein RS, Sen GC, Diamond MS. Szretter KJ, et al. PLoS Pathog. 2012;8(5):e1002698. doi: 10.1371/journal.ppat.1002698. Epub 2012 May 10. PLoS Pathog. 2012. PMID: 22589727 Free PMC article. - IFIT1: A dual sensor and effector molecule that detects non-2'-O methylated viral RNA and inhibits its translation.
Diamond MS. Diamond MS. Cytokine Growth Factor Rev. 2014 Oct;25(5):543-50. doi: 10.1016/j.cytogfr.2014.05.002. Epub 2014 May 17. Cytokine Growth Factor Rev. 2014. PMID: 24909568 Free PMC article. Review. - When your cap matters: structural insights into self vs non-self recognition of 5' RNA by immunomodulatory host proteins.
Leung DW, Amarasinghe GK. Leung DW, et al. Curr Opin Struct Biol. 2016 Feb;36:133-41. doi: 10.1016/j.sbi.2016.02.001. Epub 2016 Feb 23. Curr Opin Struct Biol. 2016. PMID: 26916433 Free PMC article. Review.
Cited by
- Application of Mammalian Nudix Enzymes to Capped RNA Analysis.
Lukaszewicz M. Lukaszewicz M. Pharmaceuticals (Basel). 2024 Sep 11;17(9):1195. doi: 10.3390/ph17091195. Pharmaceuticals (Basel). 2024. PMID: 39338357 Free PMC article. Review. - Marine fungal diversity unlocks potent antivirals against monkeypox through methyltransferase inhibition revealed by molecular dynamics and free energy landscape.
Alharbi AS, Altwaim SA, El-Daly MM, Hassan AM, Al-Zahrani IA, Bajrai LH, Alsaady IM, Dwivedi VD, Azhar EI. Alharbi AS, et al. BMC Chem. 2024 Jul 30;18(1):141. doi: 10.1186/s13065-024-01251-x. BMC Chem. 2024. PMID: 39080756 Free PMC article. - Cap-related modifications of RNA regulate binding to IFIT proteins.
Geng J, Chrabaszczewska M, Kurpiejewski K, Stankiewicz-Drogon A, Jankowska-Anyszka M, Darzynkiewicz E, Grzela R. Geng J, et al. RNA. 2024 Sep 16;30(10):1292-1305. doi: 10.1261/rna.080011.124. RNA. 2024. PMID: 39009378 Free PMC article. - Adenosine modifications impede SARS-CoV-2 RNA-dependent RNA transcription.
Snyder LR, Kilde I, Nemudryi A, Wiedenheft B, Koutmos M, Koutmou KS. Snyder LR, et al. RNA. 2024 Aug 16;30(9):1141-1150. doi: 10.1261/rna.079991.124. RNA. 2024. PMID: 38942480 Free PMC article. - 2'-_O_-methylation (Nm) in RNA: progress, challenges, and future directions.
Zhou KI, Pecot CV, Holley CL. Zhou KI, et al. RNA. 2024 Apr 16;30(5):570-582. doi: 10.1261/rna.079970.124. RNA. 2024. PMID: 38531653 Free PMC article. Review.
References
- Assenberg R., Ren J., Verma A., Walter T.S., Alderton D., Hurrelbrink R.J., Fuller S.D., Bressanelli S., Owens R.J., Stuart D.I., Grimes J.M. Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues. J. Gen. Virol. 2007;88:2228–2236. - PubMed
- Bentley D.L. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005;17:251–256. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI104002/AI/NIAID NIH HHS/United States
- R01 AI104972/AI/NIAID NIH HHS/United States
- U19 AI083019/AI/NIAID NIH HHS/United States
- U19 AI106772/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous