An estimate of the average number of recessive lethal mutations carried by humans - PubMed (original) (raw)
An estimate of the average number of recessive lethal mutations carried by humans
Ziyue Gao et al. Genetics. 2015 Apr.
Abstract
The effects of inbreeding on human health depend critically on the number and severity of recessive, deleterious mutations carried by individuals. In humans, existing estimates of these quantities are based on comparisons between consanguineous and nonconsanguineous couples, an approach that confounds socioeconomic and genetic effects of inbreeding. To overcome this limitation, we focused on a founder population that practices a communal lifestyle, for which there is almost complete Mendelian disease ascertainment and a known pedigree. Focusing on recessive lethal diseases and simulating allele transmissions, we estimated that each haploid set of human autosomes carries on average 0.29 (95% credible interval [0.10, 0.84]) recessive alleles that lead to complete sterility or death by reproductive age when homozygous. Comparison to existing estimates in humans suggests that a substantial fraction of the total burden imposed by recessive deleterious variants is due to single mutations that lead to sterility or death between birth and reproductive age. In turn, comparison to estimates from other eukaryotes points to a surprising constancy of the average number of recessive lethal mutations across organisms with markedly different genome sizes.
Keywords: autosomal recessive disease; consanguinity; human; inbreeding; recessive lethal mutation.
Copyright © 2015 by the Genetics Society of America.
Figures
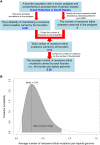
Figure 1
The analysis pipeline for estimating the average number of recessive lethal mutations carried by humans. (A) A schematic diagram of the approach. The analysis procedures are described in black. Specific values and estimates for the Hutterites are provided in blue, including the point estimate of the mean number of mutations carried by a founder. (B) The posterior distribution of the mean number of recessive lethal mutations carried by each haploid human genome, given the probability of manifestation and the number of diseases observed.
Similar articles
- Evidence of the phenotypic expression of a lethal recessive allele under inbreeding in a wild population of conservation concern.
Trask AE, Bignal EM, McCracken DI, Monaghan P, Piertney SB, Reid JM. Trask AE, et al. J Anim Ecol. 2016 Jul;85(4):879-91. doi: 10.1111/1365-2656.12503. Epub 2016 Mar 21. J Anim Ecol. 2016. PMID: 26996516 - The population genetics of human disease: The case of recessive, lethal mutations.
Amorim CEG, Gao Z, Baker Z, Diesel JF, Simons YB, Haque IS, Pickrell J, Przeworski M. Amorim CEG, et al. PLoS Genet. 2017 Sep 28;13(9):e1006915. doi: 10.1371/journal.pgen.1006915. eCollection 2017 Sep. PLoS Genet. 2017. PMID: 28957316 Free PMC article. - Balancing selection on a recessive lethal deletion with pleiotropic effects on two neighboring genes in the porcine genome.
Derks MFL, Lopes MS, Bosse M, Madsen O, Dibbits B, Harlizius B, Groenen MAM, Megens HJ. Derks MFL, et al. PLoS Genet. 2018 Sep 19;14(9):e1007661. doi: 10.1371/journal.pgen.1007661. eCollection 2018 Sep. PLoS Genet. 2018. PMID: 30231021 Free PMC article. - Inferring purging from pedigree data.
Gulisija D, Crow JF. Gulisija D, et al. Evolution. 2007 May;61(5):1043-51. doi: 10.1111/j.1558-5646.2007.00088.x. Evolution. 2007. PMID: 17492959 Review. - Discovery of rare homozygous mutations from studies of consanguineous pedigrees.
Alkuraya FS. Alkuraya FS. Curr Protoc Hum Genet. 2012 Oct;Chapter 6:Unit6.12. doi: 10.1002/0471142905.hg0612s75. Curr Protoc Hum Genet. 2012. PMID: 23074070 Review.
Cited by
- Loss of function mutations in essential genes cause embryonic lethality in pigs.
Derks MFL, Gjuvsland AB, Bosse M, Lopes MS, van Son M, Harlizius B, Tan BF, Hamland H, Grindflek E, Groenen MAM, Megens HJ. Derks MFL, et al. PLoS Genet. 2019 Mar 15;15(3):e1008055. doi: 10.1371/journal.pgen.1008055. eCollection 2019 Mar. PLoS Genet. 2019. PMID: 30875370 Free PMC article. - The crucial role of genome-wide genetic variation in conservation.
Kardos M, Armstrong EE, Fitzpatrick SW, Hauser S, Hedrick PW, Miller JM, Tallmon DA, Funk WC. Kardos M, et al. Proc Natl Acad Sci U S A. 2021 Nov 30;118(48):e2104642118. doi: 10.1073/pnas.2104642118. Proc Natl Acad Sci U S A. 2021. PMID: 34772759 Free PMC article. - Adaptation, ancestral variation and gene flow in a 'Sky Island' Drosophila species.
Hill T, Unckless RL. Hill T, et al. Mol Ecol. 2021 Jan;30(1):83-99. doi: 10.1111/mec.15701. Epub 2020 Nov 7. Mol Ecol. 2021. PMID: 33089581 Free PMC article. - The critically endangered vaquita is not doomed to extinction by inbreeding depression.
Robinson JA, Kyriazis CC, Nigenda-Morales SF, Beichman AC, Rojas-Bracho L, Robertson KM, Fontaine MC, Wayne RK, Lohmueller KE, Taylor BL, Morin PA. Robinson JA, et al. Science. 2022 May 6;376(6593):635-639. doi: 10.1126/science.abm1742. Epub 2022 May 5. Science. 2022. PMID: 35511971 Free PMC article. - Patterns of reproduction and autozygosity distinguish the breeding from nonbreeding gray wolves of Yellowstone National Park.
vonHoldt BM, DeCandia AL, Cassidy KA, Stahler EE, Sinsheimer JS, Smith DW, Stahler DR. vonHoldt BM, et al. J Hered. 2024 Jul 10;115(4):327-338. doi: 10.1093/jhered/esad062. J Hered. 2024. PMID: 37793153 Free PMC article.
References
- Alexander R. P., Fang G., Rozowsky J., Snyder M., Gerstein M. B., 2010. Annotating non-coding regions of the genome. Nat. Rev. Genet. 11: 559–571. - PubMed
- Balick, D. J., R. Do, D. Reich and S. Sunyeav, 2014 Response to a population bottleneck can be used to infer recessive selection. arXiv: 1312.3028.
- Bittles A. H., Black M. L., 2010b The impact of consanguinity on neonatal and infant health. Early Hum. Dev. 86: 737–741. - PubMed
- Bittles A. H., Makov U. E., 1985. Linear regressions in the calculation of lethal gene equivalents in man. Ann. Hum. Biol. 12: 287–289. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 HD021244/HD/NICHD NIH HHS/United States
- R01 HL085197/HL/NHLBI NIH HHS/United States
- UL1 TR000430/TR/NCATS NIH HHS/United States
- R01 HD21244/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources