Bacterial Adaptation during Chronic Respiratory Infections - PubMed (original) (raw)
Review
Bacterial Adaptation during Chronic Respiratory Infections
Louise Cullen et al. Pathogens. 2015.
Abstract
Chronic lung infections are associated with increased morbidity and mortality for individuals with underlying respiratory conditions such as cystic fibrosis (CF) and chronic obstructive pulmonary disease (COPD). The process of chronic colonisation allows pathogens to adapt over time to cope with changing selection pressures, co-infecting species and antimicrobial therapies. These adaptations can occur due to environmental pressures in the lung such as inflammatory responses, hypoxia, nutrient deficiency, osmolarity, low pH and antibiotic therapies. Phenotypic adaptations in bacterial pathogens from acute to chronic infection include, but are not limited to, antibiotic resistance, exopolysaccharide production (mucoidy), loss in motility, formation of small colony variants, increased mutation rate, quorum sensing and altered production of virulence factors associated with chronic infection. The evolution of Pseudomonas aeruginosa during chronic lung infection has been widely studied. More recently, the adaptations that other chronically colonising respiratory pathogens, including Staphylococcus aureus, Burkholderia cepacia complex and Haemophilus influenzae undergo during chronic infection have also been investigated. This review aims to examine the adaptations utilised by different bacterial pathogens to aid in their evolution from acute to chronic pathogens of the immunocompromised lung including CF and COPD.
Figures
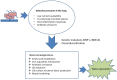
Figure 1
Selection and adaptation. Examples of selective pressures to which chronically colonising respiratory pathogens are exposed and the adaptations that they undergo, in order to enhance chances of survival.
Similar articles
- The OmpR Regulator of Burkholderia multivorans Controls Mucoid-to-Nonmucoid Transition and Other Cell Envelope Properties Associated with Persistence in the Cystic Fibrosis Lung.
Silva IN, Pessoa FD, Ramires MJ, Santos MR, Becker JD, Cooper VS, Moreira LM. Silva IN, et al. J Bacteriol. 2018 Aug 10;200(17):e00216-18. doi: 10.1128/JB.00216-18. Print 2018 Sep 1. J Bacteriol. 2018. PMID: 29914989 Free PMC article. - Genetic adaptation of Pseudomonas aeruginosa during chronic lung infection of patients with cystic fibrosis: strong and weak mutators with heterogeneous genetic backgrounds emerge in mucA and/or lasR mutants.
Ciofu O, Mandsberg LF, Bjarnsholt T, Wassermann T, Høiby N. Ciofu O, et al. Microbiology (Reading). 2010 Apr;156(Pt 4):1108-1119. doi: 10.1099/mic.0.033993-0. Epub 2009 Dec 17. Microbiology (Reading). 2010. PMID: 20019078 - Hypermutation in Burkholderia cepacia complex is mediated by DNA mismatch repair inactivation and is highly prevalent in cystic fibrosis chronic respiratory infection.
Martina P, Feliziani S, Juan C, Bettiol M, Gatti B, Yantorno O, Smania AM, Oliver A, Bosch A. Martina P, et al. Int J Med Microbiol. 2014 Nov;304(8):1182-91. doi: 10.1016/j.ijmm.2014.08.011. Epub 2014 Aug 28. Int J Med Microbiol. 2014. PMID: 25217078 - Differential adaptation of microbial pathogens to airways of patients with cystic fibrosis and chronic obstructive pulmonary disease.
Döring G, Parameswaran IG, Murphy TF. Döring G, et al. FEMS Microbiol Rev. 2011 Jan;35(1):124-46. doi: 10.1111/j.1574-6976.2010.00237.x. FEMS Microbiol Rev. 2011. PMID: 20584083 Review. - Microevolution of Pseudomonas aeruginosa to a chronic pathogen of the cystic fibrosis lung.
Hogardt M, Heesemann J. Hogardt M, et al. Curr Top Microbiol Immunol. 2013;358:91-118. doi: 10.1007/82_2011_199. Curr Top Microbiol Immunol. 2013. PMID: 22311171 Review.
Cited by
- Comparative genomics of clinical Stenotrophomonas maltophilia isolates reveals genetic diversity which correlates with colonization and persistence in vivo.
McDaniel MS, Sumpter NA, Lindgren NR, Billiot CE, Swords WE. McDaniel MS, et al. Microbiology (Reading). 2023 Nov;169(11):001408. doi: 10.1099/mic.0.001408. Microbiology (Reading). 2023. PMID: 37942787 Free PMC article. - The multifaceted role of c-di-AMP signaling in the regulation of Porphyromonas gingivalis lipopolysaccharide structure and function.
Ghods S, Muszyński A, Yang H, Seelan RS, Mohammadi A, Hilson JS, Keiser G, Nichols FC, Azadi P, Ernst RK, Moradali F. Ghods S, et al. Front Cell Infect Microbiol. 2024 Jun 12;14:1418651. doi: 10.3389/fcimb.2024.1418651. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38933693 Free PMC article. - Variation of Burkholderia cenocepacia cell wall morphology and mechanical properties during cystic fibrosis lung infection, assessed by atomic force microscopy.
Hassan AA, Vitorino MV, Robalo T, Rodrigues MS, Sá-Correia I. Hassan AA, et al. Sci Rep. 2019 Nov 6;9(1):16118. doi: 10.1038/s41598-019-52604-9. Sci Rep. 2019. PMID: 31695169 Free PMC article. - Understanding the Entanglement: Neutrophil Extracellular Traps (NETs) in Cystic Fibrosis.
Martínez-Alemán SR, Campos-García L, Palma-Nicolas JP, Hernández-Bello R, González GM, Sánchez-González A. Martínez-Alemán SR, et al. Front Cell Infect Microbiol. 2017 Apr 6;7:104. doi: 10.3389/fcimb.2017.00104. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28428948 Free PMC article. Review. - Experimental evolution and bacterial resistance: (co)evolutionary costs and trade-offs as opportunities in phage therapy research.
Scanlan PD, Buckling A, Hall AR. Scanlan PD, et al. Bacteriophage. 2015 May 21;5(2):e1050153. doi: 10.1080/21597081.2015.1050153. eCollection 2015 Apr-Jun. Bacteriophage. 2015. PMID: 26459626 Free PMC article.
References
- Zakharkina T., Heinzel E., Koczulla R.A., Greulich T., Rentz K., Pauling J.K., Baumbach J., Herrmann M., Grunewald C., Dienemann H. Analysis of the airway microbiota of healthy individuals and patients with chronic obstructive pulmonary disease by t-rflp and clone sequencing. PLoS One. 2013;8:e68302. doi: 10.1371/journal.pone.0068302. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources