Nuclear pores. Architecture of the nuclear pore complex coat - PubMed (original) (raw)
Nuclear pores. Architecture of the nuclear pore complex coat
Tobias Stuwe et al. Science. 2015.
Abstract
The nuclear pore complex (NPC) constitutes the sole gateway for bidirectional nucleocytoplasmic transport. Despite half a century of structural characterization, the architecture of the NPC remains unknown. Here we present the crystal structure of a reconstituted ~400-kilodalton coat nucleoporin complex (CNC) from Saccharomyces cerevisiae at a 7.4 angstrom resolution. The crystal structure revealed a curved Y-shaped architecture and the molecular details of the coat nucleoporin interactions forming the central "triskelion" of the Y. A structural comparison of the yeast CNC with an electron microscopy reconstruction of its human counterpart suggested the evolutionary conservation of the elucidated architecture. Moreover, 32 copies of the CNC crystal structure docked readily into a cryoelectron tomographic reconstruction of the fully assembled human NPC, thereby accounting for ~16 megadalton of its mass.
Copyright © 2015, American Association for the Advancement of Science.
Figures
Fig. 1. Overall architecture of the CNC
(A) Domain structures of the yeast coat nups and sAB-57. Black lines indicate the crystallized fragments. U: unstructured, D: domain invasion motif, VH: heavy chain variable region, CH: heavy chain constant region, VL: light chain variable region, CL: light chain constant region. (B) Reconstitution of the yeast CNC•sAB-57, lacking Nup133. Elution profiles from a Superdex 200 10/300 column are shown for Nup120•Seh1•Nup85 (Trimer 1), Sec13•Nup145C•Nup84NTD (Trimer 2), CNC, and CNC•sAB-57 (left). SDS-PAGE gel of the reconstituted CNC•sAB-57 used for crystallization (right). (C) Cartoon and schematic representations of the yeast CNC•sAB-57 crystal structure viewed from two sides.
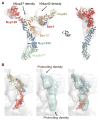
Fig. 2. Architecture of the CNC triskelion
Cartoon representation of the triskelion formed by Nup120, Nup85 and Nup145C. Insets (A–C) depict magnified views for the interactions between (A) Nup120CTD, Nup85CTD, and Nup145CCTD (B) Nup120CTD, Nup85CTD, and N-terminal Nup145C helix; and (C) Nup145C, Nup85CTD, and sAB-57. The density modified electron density map is contoured at 1.0 σ.
Fig. 3. Comparison of yeast and human CNCs
(A) Fit of the yeast CNC crystal structure into the human CNC negative-stain EM reconstruction (gray) (3). Arrows indicate density accounted for by the additional human coat nups Nup37 or Nup43. (B) Comparison of the quality of fit for the yeast CNC crystal structure and human CNC EM reconstruction (cyan) into the intact human NPC cryoelectron tomographic reconstruction (gray) (3). Arrows indicate regions where the human CNC EM reconstruction protrudes from the cryoelectron tomographic reconstruction.
Fig. 4. Architecture of the NPC coat
(A) 32 copies of the yeast CNC, shown in cartoon representation with a representative subunit colored as in Fig. 1, docked into the cryoelectron tomographic reconstruction of the intact human NPC (3), shown as a gray surface. The outer and inner cytoplasmic and nuclear CNC rings are highlighted in orange, cyan, pink, and blue, respectively. (B) Cartoon representations of 16 yeast CNC copies from the cytoplasmic side of the NPC coat. Schematics indicating the positions assigned to Nup84CTD and Nup133, which were not crystallized, are shown. (C) Interface between the inner and outer CNC rings. Two views of the yeast CNC and its mate from the inner ring are shown. (D) Orientation of the Nup120 β-propeller relative to neighboring coat nups and the membrane. Portions of two CNCs from the cytoplasmic outer ring are shown in cartoon representation. Green and cyan shading indicates the positioning of Nup84CTD and Nup133, respectively. The cyan line represents the N-terminal unstructured segment of Nup133 that binds to Nup120 (9). A schematic representation of the ring-forming Nup120-Nup133 interaction is shown below.
Similar articles
- Architecture of the linker-scaffold in the nuclear pore.
Petrovic S, Samanta D, Perriches T, Bley CJ, Thierbach K, Brown B, Nie S, Mobbs GW, Stevens TA, Liu X, Tomaleri GP, Schaus L, Hoelz A. Petrovic S, et al. Science. 2022 Jun 10;376(6598):eabm9798. doi: 10.1126/science.abm9798. Epub 2022 Jun 10. Science. 2022. PMID: 35679425 Free PMC article. - Architecture of the cytoplasmic face of the nuclear pore.
Bley CJ, Nie S, Mobbs GW, Petrovic S, Gres AT, Liu X, Mukherjee S, Harvey S, Huber FM, Lin DH, Brown B, Tang AW, Rundlet EJ, Correia AR, Chen S, Regmi SG, Stevens TA, Jette CA, Dasso M, Patke A, Palazzo AF, Kossiakoff AA, Hoelz A. Bley CJ, et al. Science. 2022 Jun 10;376(6598):eabm9129. doi: 10.1126/science.abm9129. Epub 2022 Jun 10. Science. 2022. PMID: 35679405 Free PMC article. - Toward the atomic structure of the nuclear pore complex: when top down meets bottom up.
Hoelz A, Glavy JS, Beck M. Hoelz A, et al. Nat Struct Mol Biol. 2016 Jul;23(7):624-30. doi: 10.1038/nsmb.3244. Epub 2016 Jun 6. Nat Struct Mol Biol. 2016. PMID: 27273515 Free PMC article. Review. - Architecture of the symmetric core of the nuclear pore.
Lin DH, Stuwe T, Schilbach S, Rundlet EJ, Perriches T, Mobbs G, Fan Y, Thierbach K, Huber FM, Collins LN, Davenport AM, Jeon YE, Hoelz A. Lin DH, et al. Science. 2016 Apr 15;352(6283):aaf1015. doi: 10.1126/science.aaf1015. Epub 2016 Apr 14. Science. 2016. PMID: 27081075 Free PMC article. - The Structure of the Nuclear Pore Complex (An Update).
Lin DH, Hoelz A. Lin DH, et al. Annu Rev Biochem. 2019 Jun 20;88:725-783. doi: 10.1146/annurev-biochem-062917-011901. Epub 2019 Mar 18. Annu Rev Biochem. 2019. PMID: 30883195 Free PMC article. Review.
Cited by
- A short linear motif in scaffold Nup145C connects Y-complex with pre-assembled outer ring Nup82 complex.
Teimer R, Kosinski J, von Appen A, Beck M, Hurt E. Teimer R, et al. Nat Commun. 2017 Oct 24;8(1):1107. doi: 10.1038/s41467-017-01160-9. Nat Commun. 2017. PMID: 29062044 Free PMC article. - Structure and drug resistance of the Plasmodium falciparum transporter PfCRT.
Kim J, Tan YZ, Wicht KJ, Erramilli SK, Dhingra SK, Okombo J, Vendome J, Hagenah LM, Giacometti SI, Warren AL, Nosol K, Roepe PD, Potter CS, Carragher B, Kossiakoff AA, Quick M, Fidock DA, Mancia F. Kim J, et al. Nature. 2019 Dec;576(7786):315-320. doi: 10.1038/s41586-019-1795-x. Epub 2019 Nov 27. Nature. 2019. PMID: 31776516 Free PMC article. - In situ architecture of the algal nuclear pore complex.
Mosalaganti S, Kosinski J, Albert S, Schaffer M, Strenkert D, Salomé PA, Merchant SS, Plitzko JM, Baumeister W, Engel BD, Beck M. Mosalaganti S, et al. Nat Commun. 2018 Jun 18;9(1):2361. doi: 10.1038/s41467-018-04739-y. Nat Commun. 2018. PMID: 29915221 Free PMC article. - Dissecting Torsin/cofactor function at the nuclear envelope: a genetic study.
Laudermilch E, Tsai PL, Graham M, Turner E, Zhao C, Schlieker C. Laudermilch E, et al. Mol Biol Cell. 2016 Dec 15;27(25):3964-3971. doi: 10.1091/mbc.E16-07-0511. Epub 2016 Oct 26. Mol Biol Cell. 2016. PMID: 27798237 Free PMC article. - Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex.
Li Z, Chen S, Zhao L, Huang G, Pi X, Sun S, Wang P, Sui SF. Li Z, et al. Cell Res. 2022 May;32(5):437-450. doi: 10.1038/s41422-022-00632-y. Epub 2022 Mar 18. Cell Res. 2022. PMID: 35301440 Free PMC article.
References
- Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–643. - PubMed
- Bui KH, et al. Integrated structural analysis of the human nuclear pore complex scaffold. Cell. 2013;155:1233–1243. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM007616/GM/NIGMS NIH HHS/United States
- U01 GM094588/GM/NIGMS NIH HHS/United States
- ACB-12002/PHS HHS/United States
- AGM-12006/PHS HHS/United States
- 5 T32 GM07616/GM/NIGMS NIH HHS/United States
- U54 GM087519/GM/NIGMS NIH HHS/United States
- R01 GM117360/GM/NIGMS NIH HHS/United States
- R01 GM111461/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases