A Dual-reporter system for real-time monitoring and high-throughput CRISPR/Cas9 library screening of the hepatitis C virus - PubMed (original) (raw)
A Dual-reporter system for real-time monitoring and high-throughput CRISPR/Cas9 library screening of the hepatitis C virus
Qingpeng Ren et al. Sci Rep. 2015.
Abstract
The hepatitis C virus (HCV) is one of the leading causes of chronic hepatitis, liver cirrhosis and hepatocellular carcinomas and infects approximately 170 million people worldwide. Although several reporter systems have been developed, many shortcomings limit their use in the assessment of HCV infections. Here, we report a real-time live-cell reporter, termed the NIrD (NS3-4A Inducible rtTA-mediated Dual-reporter) system, which provides an on-off switch specifically in response to an HCV infection. Using the NIrD system and a focused CRISPR/Cas9 library, we identified CLDN1, OCLN and CD81 as essential genes for both the cell-free entry and the cell-to-cell transmission of HCV. The combination of this ultra-sensitive reporter system and the CRISPR knockout screening provides a powerful and high-throughput strategy for the identification of critical host components for HCV infections.
Figures
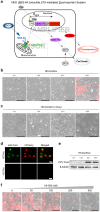
Figure 1. A cell-based dual-reporter system for monitoring HCV infections.
(a) The rationale of the NIrD system. The sensor module consists of rtTA-MAVS(C) fusion proteins, which are constantly produced and localised to the mitochondria in cells. The amplifier module is an expression cartridge integrated into the chromosome that is composed of two reporter genes driven by the tight-TRE promoter. Upon HCV infection, the virally produced NS3-4A cleaves its recognition sequence in MAVS(C), releasing the free-formed rtTA into the nucleus. The tight-TRE promoter is activated by rtTA and Dox (2 μg/ml), resulting in the production of delta-TK-2A-mCherry proteins. After self-cleavage of the 2A peptide, mCherry shows red fluorescence and delta-TK phosphorylates GCV (2 μg/ml) to cause cell death. (b) mCherry signal of the NIrD system in response to an HCVcc infection. The Huh7.5(NIrD) system was infected by HCVcc for 72 h. HCVcc-transduced (+) or -untransduced (−) cells in the presence (+) or absence (−) of Dox (2 μg/ml) were visualised under a microscope. The fluorescence signals were superimposed onto white light images. Scale bar, 200 μm. (c) The death signal of the NIrD system in response to an HCVcc infection in the presence of GCV (2 μg/ml). Huh7.5(NIrD) cells were infected with HCVcc for 120 h. The transduced (+) or untransduced (−) cells in the presence (+) or absence (−) of Dox (2 μg/ml) were visualised under a light microscope. Scale bar, 200 μm. (d) Fluorescence microscopy of HCVcc core protein (Alexa Fluor 488) and mCherry signal upon HCVcc infection (72 h) in Huh7.5(NIrD) cells. Scale bar, 30 μm. (e) Immunoblotting analysis of Huh7.5(NIrD) cells infected with or without HCVcc in the presence or absence of Dox (2 μg/ml). HCVcc was detected by an antibody specifically targeting the viral core protein, and β-tubulin was used as the loading control. (f) Dosage effects of VX-950 on the live-cell imaging of HCVcc-infected Huh7.5(NIrD) cells. Huh7.5(NIrD) cells were infected with HCVcc plus Dox (2 μg/ml) together with serially increasing dosages of VX-950 (Telaprevir). Fluorescence microscopic images were taken 72 h following the HCVcc infection. Scale bar, 200 μm.
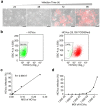
Figure 2. Quantitative evaluation of NIrD system in response to HCV inoculation.
(a) Time-lapse live-cell imaging of Huh7.5(NIrD) cells. Both light and fluorescence images were taken every 24 h, starting at 24 h post-HCVcc infection in the presence of Dox (2 μg/ml). Scale bar, 100 μm. (b) FACS analysis of Huh7.5(NIrD) cells infected by HCVcc in the presence of Dox (2 μg/ml). A total of 2 × 105/well of Huh7.5(NIrD) cells were seeded in 6-well plates. Representative results from reporter cells treated with HCVcc (0 or 25,100 TCID50/ml) are presented. FACS analysis was conducted 96 h following the viral infection. The numbers in the square indicate the percentage of red fluorescence-negative cells. (c-d) FACS analysis of Huh7.5(NIrD) cells infected by serially increasing dosages of HCVcc. The curves show the percentage of mCherry positive cells corresponding to MOI (2 × 105 cells/well) in linear (c) or logarithmic (d) plots.
Figure 3. Schematic of the CRISPR library construction and HCV screening.
(a) The structure of the lentiviral plasmid expressing OCT1 and Cas9. (b) Indels induced by the lentivirus-delivered sgRNA (5-TTGGCCAGACTTGCATCCG-3) targeting the CSPG4 gene in the indicated cells were assayed by T7E1 digestion. Genomic DNA from HeLaOC-SC was used as a positive control, and the wild type (WT) Huh7.5(NIrD) was used as a negative control. (c) Schematic of the sgRNA library screening. sgRNAs were delivered into Huh7.5(NIrD)OC-SC cells by lentiviral infection with a MOI of 0.1. Three replicates of the libraries were challenged with 3–4 rounds of HCVcc, followed by FACS sorting to enrich the mCherry-negative clones. A comparison of the abundance of sgRNAs between the treated and untreated populations through high-throughput sequencing analysis was conducted following the same procedure as previously reported.
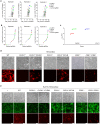
Figure 4. Screen of host genes essential for an HCV infection.
(a) Primary HCVcc screening data. The count of every sgRNA is the number of reads that match the sgRNA target sequence. (b) sgRNA ranking was based on the fold change of normalised counts of every sgRNA in the HCV treated and untreated populations. (c) The FDR (false discovery rate) of every gene in the library was calculated by MAGeCK based on the counts and kinds of sgRNAs in the three replicates. (d) Effects of the gene knockout of CLND1, OCLN and CD81 on cell-free entry of HCVcc. All cells indicated carry the NIrD system. Light and fluorescence images were taken 72 h post-HCVcc infection in the presence of Dox (2 μg/ml). Scale bar, 100 μm. (e) Effects of the gene knockout of CLND1, OCLN and CD81 on cell-to-cell transmission of HCVcc. Huh7.5(NΙrD) cells with the indicated background (WT, _CLDN1_−/−, _CLDN1_−/−/CLDN1, _OCLN_−/−, _OCLN_−/−/OCLN, _CD81_−/− or _CD81_−/−/CD81) were co-cultured with HCVcc pre-infected (24 h prior) Huh7.5 cells. HCVcc carries the EGFP gene in its genome, resulting in a punctuated green fluorescence pattern in the cells. _OCLN_−/− and _CD81_−/− knockout cells expressed diffused green fluorescence because they were derived from cells expressing EGFP. The light and fluorescence (green and red) images were taken 72 h following the co-culturing of the cells. Scale bar, 100 μm.
Similar articles
- Attachment and Postattachment Receptors Important for Hepatitis C Virus Infection and Cell-to-Cell Transmission.
Fan H, Qiao L, Kang KD, Fan J, Wei W, Luo G. Fan H, et al. J Virol. 2017 Jun 9;91(13):e00280-17. doi: 10.1128/JVI.00280-17. Print 2017 Jul 1. J Virol. 2017. PMID: 28404852 Free PMC article. - Visualizing the Essential Role of Complete Virion Assembly Machinery in Efficient Hepatitis C Virus Cell-to-Cell Transmission by a Viral Infection-Activated Split-Intein-Mediated Reporter System.
Zhao F, Zhao T, Deng L, Lv D, Zhang X, Pan X, Xu J, Long G. Zhao F, et al. J Virol. 2017 Jan 3;91(2):e01720-16. doi: 10.1128/JVI.01720-16. Print 2017 Jan 15. J Virol. 2017. PMID: 27852847 Free PMC article. - Mice Expressing Minimally Humanized CD81 and Occludin Genes Support Hepatitis C Virus Uptake In Vivo.
Ding Q, von Schaewen M, Hrebikova G, Heller B, Sandmann L, Plaas M, Ploss A. Ding Q, et al. J Virol. 2017 Jan 31;91(4):e01799-16. doi: 10.1128/JVI.01799-16. Print 2017 Feb 15. J Virol. 2017. PMID: 27928007 Free PMC article. - Advances with using CRISPR/Cas-mediated gene editing to treat infections with hepatitis B virus and hepatitis C virus.
Moyo B, Bloom K, Scott T, Ely A, Arbuthnot P. Moyo B, et al. Virus Res. 2018 Jan 15;244:311-320. doi: 10.1016/j.virusres.2017.01.003. Epub 2017 Jan 10. Virus Res. 2018. PMID: 28087399 Review. - High-throughput screens in mammalian cells using the CRISPR-Cas9 system.
Peng J, Zhou Y, Zhu S, Wei W. Peng J, et al. FEBS J. 2015 Jun;282(11):2089-96. doi: 10.1111/febs.13251. Epub 2015 Mar 16. FEBS J. 2015. PMID: 25731961 Review.
Cited by
- A genome-wide CRISPR screening uncovers that TOB1 acts as a key host factor for FMDV infection via both IFN and EGFR mediated pathways.
Peng G, Liu T, Qi X, Wang Y, Ren J, Peng J, Du X, Hu S, Wu S, Zhao Y, Li D, Zheng H. Peng G, et al. PLoS Pathog. 2024 Mar 21;20(3):e1012104. doi: 10.1371/journal.ppat.1012104. eCollection 2024 Mar. PLoS Pathog. 2024. PMID: 38512977 Free PMC article. - A genome-wide CRISPR/Cas9 screen identifies a role for Rab5A and early endosomes in hepatitis E virus replication.
Oechslin N, Da Silva N, Ankavay M, Moradpour D, Gouttenoire J. Oechslin N, et al. Proc Natl Acad Sci U S A. 2023 Dec 26;120(52):e2307423120. doi: 10.1073/pnas.2307423120. Epub 2023 Dec 18. Proc Natl Acad Sci U S A. 2023. PMID: 38109552 Free PMC article. - CRISPR-Cas system to discover host-virus interactions in Flaviviridae.
Ramezannia Z, Shamekh A, Bannazadeh Baghi H. Ramezannia Z, et al. Virol J. 2023 Oct 27;20(1):247. doi: 10.1186/s12985-023-02216-7. Virol J. 2023. PMID: 37891676 Free PMC article. Review. - The non-classical major histocompatibility complex II protein SLA-DM is crucial for African swine fever virus replication.
Pannhorst K, Carlson J, Hölper JE, Grey F, Baillie JK, Höper D, Wöhnke E, Franzke K, Karger A, Fuchs W, Mettenleiter TC. Pannhorst K, et al. Sci Rep. 2023 Aug 21;13(1):10342. doi: 10.1038/s41598-023-36788-9. Sci Rep. 2023. PMID: 37604847 Free PMC article. - Multi-faceted CRISPR/Cas technological innovation aspects in the framework of 3P medicine.
Lučanský V, Holubeková V, Kolková Z, Halašová E, Samec M, Golubnitschaja O. Lučanský V, et al. EPMA J. 2023 May 22;14(2):201-217. doi: 10.1007/s13167-023-00324-6. eCollection 2023 Jun. EPMA J. 2023. PMID: 37275547 Free PMC article. Review.
References
- Choo Q. L. et al. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244, 359–362 (1989). - PubMed
- Andreoni M. et al. HIV-HCV co-infection: epidemiology, pathogenesis and therapeutic implications. Eur Rev Med Pharmacol Sci 16, 1473–1483 (2012). - PubMed
- Moradpour D., Penin F. & Rice C. M. Replication of hepatitis C virus. Nat Rev Microbiol 5, 453–463 (2007). - PubMed
- Pileri P. et al. Binding of hepatitis C virus to CD81. Science 282, 938–941 (1998). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous