An allosteric model for control of pore opening by substrate binding in the EutL microcompartment shell protein - PubMed (original) (raw)
An allosteric model for control of pore opening by substrate binding in the EutL microcompartment shell protein
Michael C Thompson et al. Protein Sci. 2015 Jun.
Abstract
The ethanolamine utilization (Eut) microcompartment is a protein-based metabolic organelle that is strongly associated with pathogenesis in bacteria that inhabit the human gut. The exterior shell of this elaborate protein complex is composed from a few thousand copies of BMC-domain shell proteins, which form a semi-permeable diffusion barrier that provides the interior enzymes with substrates and cofactors while simultaneously retaining metabolic intermediates. The ability of this protein shell to regulate passage of substrate and cofactor molecules is critical for microcompartment function, but the details of how this diffusion barrier can allow the passage of large cofactors while still retaining small intermediates remain unclear. Previous work has revealed two conformations of the EutL shell protein, providing substantial evidence for a gated pore that might allow the passage of large cofactors. Here we report structural and biophysical evidence to show that ethanolamine, the substrate of the Eut microcompartment, acts as a negative allosteric regulator of EutL pore opening. Specifically, a series of X-ray crystal structures of EutL from Clostridium perfringens, along with equilibrium binding studies, reveal that ethanolamine binds to EutL at a site that exists in the closed-pore conformation and which is incompatible with opening of the large pore for cofactor transport. The allosteric mechanism we propose is consistent with the cofactor requirements of the Eut microcompartment, leading to a new model for EutL function. Furthermore, our results suggest the possibility of redox modulation of the allosteric mechanism, opening potentially new lines of investigation.
Keywords: X-ray crystallography; allostery; bacterial microcompartment; conformational change; ethanolamine metabolism; ligand-binding.
© 2015 The Protein Society.
Figures
Figure 1
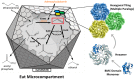
The Eut MCP is a bacterial organelle that functions to retain acetaldehyde (red box), a volatile and toxic metabolic intermediate. It consists of BMC-domain proteins, hexagonally tiled to form a polyhedral shell around a series of sequentially-acting enzymes that convert ethanolamine to ethanol and acetyl phosphate. Primarily small molecules, substrates and products, must traverse the shell, while the NAD(H) and acetyl CoA cofactors are recycled within the MCP. In contrast, the cobalamin cofactor (orange) required by the EutBC enzyme must be regenerated under certain conditions, requiring exchange of a large molecule between the MCP lumen and the cytosol.
Figure 2
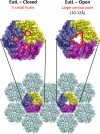
The closed and open structures of EutL, as established from previous studies., The comparison shown is of EutL from E. coli, which is the only homolog whose structure has been determined in both open and closed forms. EutL is shown in a hexagonal tiling with EutM hexamers (light blue). The expanded views show key features of the two conformations, including three narrow holes (channels) in the closed conformation, and a large central pore in the open conformation. The individual chains of the EutL pseudohexameric trimer are each colored differently (purple, yellow, dark blue).
Figure 3
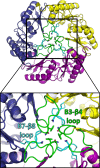
Crystal structure of EutL from Clostridium perfringens (CpEutL)(this work), colored as in Figure 2 but with the β3–β4 loops colored green, and the β7–β8 loops colored cyan. Three symmetry-related copies of each of these two loops pack tightly at the center of the trimer, resulting in a conformation that is essentially closed to transport. Three conserved amino acids that appear to be important for stabilizing this closed conformation (Y69, N74, N183) are shown as sticks.
Figure 4
Visualization of narrow holes in the closed conformation of CpEutL. (A) Each CpEutL trimer presents three small holes, one in each monomer, which are also present in the closed structure of E. coli EutL. The gray surface serves to highlight these holes or channels, while the cyan spheres denote the Cα atoms of the residues that form the narrowest constriction points (D44, D45, V151, T182, and F184). (B) The open and closed structures of E. coli EutL demonstrate that opening of the central pore involves retraction of the central loops (magenta), into the small channels (cyan spheres). (C) Analysis of the narrow channels using the HOLE2 algorithm reveals that their narrowest constriction point has a radius of only 1.3Å.
Figure 5
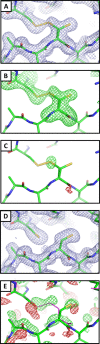
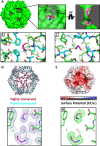
Visualization of a disulfide bond in CpEutL. (A) A 2mFo − DFc electron density map contoured at 1.0_σ_ shows contiguous electron density between the sulfur atoms of C127 and C200, indicating the presence of a partially occupied disulfide bond. (B) An omit (mFo − DFc, 3.0_σ_) electron density map, in which the atoms of Cys127 and Cys200 were excluded from the structure factor and phase calculations to eliminate model bias, shows strong positive density for both conformations of Cys127 and for the disulfide bond. (C) An _F_early − F_late (φ_model, 4.0_σ), electron density map, comparing data from the first 20° of Xray data collection with the last 20°, indicates that the Cys127–Cys200 disulfide bond is strongly populated prior to X-ray exposure, and is partially destroyed during the experiment. (D) Chemical reduction abolishes the disulfide bond in CpEutL, as evidenced by a lack of electron density between the cysteine side chains in the 2mFo − DFc (1.0_σ) electron density map derived from the reduced crystals. (E) An isomorphous difference map calculated using _F_oxidized − _F_reduced amplitudes (φ_model, 3.0_σ) show positive peaks between the sulfur atoms of C127 and C200, further verifying the existence of the disulfide bond. Experimental data (see text) shows that the disulfide bond is linked to substrate binding and allosteric modulation of the large central pore in EutL.
Figure 6
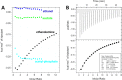
Thermodynamic characterization of ethanolamine binding to EutL. (A) A comparison of integrated ITC data for titrations of several small molecules into a CpEutL solution reveals that ethanolamine is the only titrant whose interaction with CpEutL releases heat in a manner consistent with a specific binding event, which appears to be highly selective for ethanolamine. (B) Plots of the raw (top panel) and integrated (bottom panel) ITC data measured for a titration of ethanolamine into CpEutL. The data shown were used to derive thermodynamic parameters for the ethanolamine-CpEutL interaction using a sequential, two-site binding model.
Figure 7
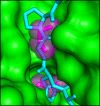
Crystallographic visualization of ethanolamine binding to EutL. (A) X-ray crystal structures reveal that ethanolamine molecules (magenta) bind to EutL in the narrow channels that perforate the trimers. Each monomer contains a single channel with two bound ethanolamine molecules. Within each channel, one ethanolamine molecule is bound on either side of the narrow, hourglass-shaped constriction point. Conserved amino acids (cyan) form hydrophobic and polar interactions with ethanolamine molecules (magenta) that occupy each of the two binding sites (panels B and C). (D) A cartoon diagram of CpEutL with Cα atoms depicted as spheres and colored according to sequence conservation scores, where dark magenta indicates highly conserved positions, and dark cyan indicates poorly conserved positions. Note the high degree of conservation among residues surrounding the ethanolamine binding sites. E) An electrostatic surface representation of CpEutL, calculated using the Poisson-Boltzmann equation, shows that the ethanolamine binding sites bear a strong negative charge, complementary to their ligand. (F) A 2mFo − DFc electron density map (φ_model, 1.0_σ) supports the placement of the ethanolamine ligands (magenta) in the crystallographic model. G) The presence of the ethanolamine ligands was confirmed by calculation of an omit (mFo − DFc, 3.0_σ_) electron density map, in which the ethanolamine molecules were excluded from the structure factor and phase calculations. This map shows exceptionally strong and well-resolved peaks corresponding to the ethanolamine molecules.
Figure 8
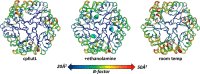
Evaluation of mobility in cryogenic and room temperature structures of EutL. Three structures of CpEutL, colored according to the average atomic B-factor for each residue, and also rendered such that the width of the cartoon diagram represents the average atomic B-factor of each residue, all show low B-factors for the ethanolamine binding channels, indicating these holes do not undergo fluctuations of sufficient amplitude to allow the passage of ethanolamine. The structure on the left is the untreated, cryogenic structure, the structure in the middle is the ethanolamine-bound, cryogenic structure, and the structure on the right is the untreated, room temperature structure.
Figure 9
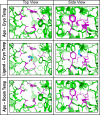
Examination of mobility based on ensemble refinement of EutL structures. The images show the ethanolamine binding channels in CpEutL as modeled using time-averaged ensemble refinement against cryogenic CpEutL diffraction data collected from both apo and ethanolaminebound (top and middle panels respectively) crystals, and also against CpEutL diffraction data collected from apo crystals at room temperature (bottom panel). The residues that form the constriction point of the hourglass-shaped channel are shown in magenta. The ensembles show that the channels do not undergo any significant dynamic expansion that might permit substrate transport.
Figure 10
A steric clash forms the basis for allosteric regulation. The cutaway view depicts the ethanolamine binding channel as a molecular surface, with the ethanolamine molecules shown as magenta sticks surrounded by transparent spheres. The cyan polypeptide shows the open conformation of E. coli EutL superimposed on the CpEutL:ethanolamine complex. The overlay reveals that ethanolamine binding prevents a rearrangement from the closed conformation to the open conformation by blocking the space into which the β3–β4 loop moves during the transition.
Figure 11
A model for allosteric regulation of cofactor transport in the Eut MCP. (A) Negative allosteric regulation of EutL pore opening is achieved by ethanolamine binding to the closed conformation of EutL, resulting in steric hindrance that prevents the transition to the open conformation. B) We propose a new model for EutL function in the context of the complex enzymology of the Eut MCP. The cobalamin cofactor of the encapsulated EutBC enzyme spontaneously inactivates over time. When the ethanolamine substrate is absent (or has been metabolized), the EutL shell protein remains in the open form so its large central pore can transport large cofactors for enzyme maintenance. Then, when ethanolamine is present, closure of the large EutL pore allows metabolism of ethanolamine to occur without release of the toxic acetaldehyde intermediate. Note that ethanolamine uptake is proposed to occur through a different shell protein paralog (EutM), whose central pore is narrow and selective, but constitutively open.
Similar articles
- Crystallographic insights into the pore structures and mechanisms of the EutL and EutM shell proteins of the ethanolamine-utilizing microcompartment of Escherichia coli.
Takenoya M, Nikolakakis K, Sagermann M. Takenoya M, et al. J Bacteriol. 2010 Nov;192(22):6056-63. doi: 10.1128/JB.00652-10. Epub 2010 Sep 17. J Bacteriol. 2010. PMID: 20851901 Free PMC article. - Structure and mechanisms of a protein-based organelle in Escherichia coli.
Tanaka S, Sawaya MR, Yeates TO. Tanaka S, et al. Science. 2010 Jan 1;327(5961):81-4. doi: 10.1126/science.1179513. Science. 2010. PMID: 20044574 - Structure of a bacterial microcompartment shell protein bound to a cobalamin cofactor.
Thompson MC, Crowley CS, Kopstein J, Bobik TA, Yeates TO. Thompson MC, et al. Acta Crystallogr F Struct Biol Commun. 2014 Dec 1;70(Pt 12):1584-90. doi: 10.1107/S2053230X1402158X. Epub 2014 Nov 14. Acta Crystallogr F Struct Biol Commun. 2014. PMID: 25484204 Free PMC article. - Diverse bacterial microcompartment organelles.
Chowdhury C, Sinha S, Chun S, Yeates TO, Bobik TA. Chowdhury C, et al. Microbiol Mol Biol Rev. 2014 Sep;78(3):438-68. doi: 10.1128/MMBR.00009-14. Microbiol Mol Biol Rev. 2014. PMID: 25184561 Free PMC article. Review. - Recent structural insights into bacterial microcompartment shells.
Ochoa JM, Yeates TO. Ochoa JM, et al. Curr Opin Microbiol. 2021 Aug;62:51-60. doi: 10.1016/j.mib.2021.04.007. Epub 2021 May 28. Curr Opin Microbiol. 2021. PMID: 34058518 Free PMC article. Review.
Cited by
- Evolutionary relationships among shell proteins of carboxysomes and metabolosomes.
Melnicki MR, Sutter M, Kerfeld CA. Melnicki MR, et al. Curr Opin Microbiol. 2021 Oct;63:1-9. doi: 10.1016/j.mib.2021.05.011. Epub 2021 Jun 5. Curr Opin Microbiol. 2021. PMID: 34098411 Free PMC article. Review. - Prokaryotic Organelles: Bacterial Microcompartments in E. coli and Salmonella.
Stewart KL, Stewart AM, Bobik TA. Stewart KL, et al. EcoSal Plus. 2020 Oct;9(1):10.1128/ecosalplus.ESP-0025-2019. doi: 10.1128/ecosalplus.ESP-0025-2019. EcoSal Plus. 2020. PMID: 33030141 Free PMC article. Review. - Monatomic ions influence substrate permeation across bacterial microcompartment shells.
Trettel DS, Neale C, Zhao M, Gnanakaran S, Gonzalez-Esquer CR. Trettel DS, et al. Sci Rep. 2023 Sep 21;13(1):15738. doi: 10.1038/s41598-023-42688-9. Sci Rep. 2023. PMID: 37735196 Free PMC article. - Selective molecular transport across the protein shells of bacterial microcompartments.
Bobik TA, Stewart AM. Bobik TA, et al. Curr Opin Microbiol. 2021 Aug;62:76-83. doi: 10.1016/j.mib.2021.05.006. Epub 2021 Jun 1. Curr Opin Microbiol. 2021. PMID: 34087617 Free PMC article. Review. - A Bacterial Microcompartment Is Used for Choline Fermentation by Escherichia coli 536.
Herring TI, Harris TN, Chowdhury C, Mohanty SK, Bobik TA. Herring TI, et al. J Bacteriol. 2018 Apr 24;200(10):e00764-17. doi: 10.1128/JB.00764-17. Print 2018 May 15. J Bacteriol. 2018. PMID: 29507086 Free PMC article.
References
- Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. - PubMed
- Guarner F, Malagelada J-R. Gut flora in health and disease. Lancet. 2003;361:512–519. - PubMed
- Sears CL. A dynamic partnership: celebrating our gut flora. Anaerobe. 2005;11:247–251. - PubMed
- Cummings JH, Macfarlane GT. Role of intestinal bacteria in nutrient metabolism. Clin Nutr. 1997;16:3–11. - PubMed
- Steinhoff U. Who controls the crowd? New findings and old questions about the intestinal microflora. Immunol Lett. 2005;99:12–16. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources