Genomic analysis of LPS-stimulated myeloid cells identifies a common pro-inflammatory response but divergent IL-10 anti-inflammatory responses - PubMed (original) (raw)
Genomic analysis of LPS-stimulated myeloid cells identifies a common pro-inflammatory response but divergent IL-10 anti-inflammatory responses
Andrew Paul Hutchins et al. Sci Rep. 2015.
Abstract
Inflammation is an essential physiological response to infection and injury that must be kept within strict bounds. The IL-10/STAT3 anti-inflammatory response (AIR) is indispensable for controlling the extent of inflammation, although the complete mechanisms downstream of STAT3 have not yet been elucidated. The AIR is widely known to extend to other myeloid cells, but it has best been characterized in macrophages. Here we set out to characterize the LPS-mediated pro-inflammatory response and the AIR across a range of myeloid cells. We found that whereas the LPS-induced pro-inflammatory response is broadly similar among macrophages, dendritic cells, neutrophils, mast cells and eosinophils, the AIR is drastically different across all myeloid cell types that respond to IL-10 (all bar eosinophils). We propose a model whereby the IL-10/STAT3 AIR works by selectively inhibiting specific pathways in distinct cell types: in macrophages the AIR most likely works through the inhibition of NF-κB target genes; in DCs and mast cells through indirect IRF disruption; and in neutrophils through IRF disruption and possibly also indirect NF-κB inhibition. In summary, no conserved IL-10/STAT3 AIR effectors were identified; instead a cell type-specific model of the AIR is proposed.
Figures
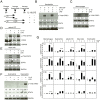
Figure 1. IL-10 leads to phosphorylation of STAT3 and activates an AIR in macrophages, neutrophils, sDC and mast cells, but not in eosinophils.
(A) Schematic of treatment scheme used in this study. Macrophages, neutrophils, splenic sDC, mast cells and eosinophils were either purified (macrophages, neutrophils, sDC) or derived (mast cells, eosinophils) from mouse tissues, treated with IL-10 for 4 h and then subsequently treated with LPS for a further 4 hours. Upon addition of IL-10 STAT3 is phosphorylated in neutrophils (B), macrophages (C), sDCs (D) and mast cells (E), but not in eosinophils (F). Full-length blots are provided in Supplementary Figure 2. (G) qRT-PCR of the pro-inflammatory cytokines Tnf (TNFa), Cxcl10 (IP10) and Il12b, which are down-regulated when IL-10 is combined with LPS treatment except in eosinophils. Error bars are 95% confidence intervals, significantly down-regulated (p < 0.05) changes between +LPS and +IL-10+LPS are indicated. Genes must first be significantly up-regulated by LPS.
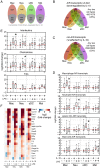
Figure 2. Principal componenet analysis of changes in myeloid gene expression.
(A) Principal component (PC) analysis of the myeloid cells assayed in this study. PCs 2 through 7 are indicated, cell types are colored according to the key and where appropriate samples that segregate with and without LPS are indicated with a line showing the separation and ‘+LPS' and ‘-LPS' for the appropriate treatment. At no PC could we detect a gradient that corresponded to IL-10 treatment. (B) Loading for PC6, which correlates with LPS treatment. (C) Heatmap of the fold-change expression changes of the top 50 genes at the top of PC6.
Figure 3. LPS endotoxin activates a potent, common pro-inflammatory response.
(A) The number of transcripts up (left) and down (right)-regulated by LPS in each cell type and in different combinations of cells. Categories are exclusive, and the total number of genes regulated in the appropriate cell type is indicated in brackets. (B) Boxplots of relative levels of expression of LPS stimulated genes specifically up-regulated in the respective cell type. (C) Venn diagrams of up and down regulated transcripts in macrophages, neutrophils and splenic DCs. (D) Gene ontology analysis for up-regulated genes in the five cell types. (E) Boxplots for all expressed interleukins, chemokines and Tnf-family members. Mann-Whitney-U test: *p-value < 0.05, **p-value < 0.01. (F) Heatmap of the cytokines/chemokines up-regulated in at least 2 cell types, ordered by overall fold-change of expression upon addition of LPS.
Figure 4. IL-10 suppression of the LPS-initiated pro-inflammatory response is divergent in the four myeloid cell types.
(A) IL-10 suppressed transcripts (AIR transcripts) were defined as those transcripts declining at least 2 fold. (B) and (C) Transcripts were divided into two categories: (i) AIR transcripts – those transcripts that decline by at least 2 fold after LPS induction in at least one cell type and (ii) ‘not AIR' transcripts that did not decline at least 2-fold after IL-10 treatment. (B) Venn diagram of the IL-10 suppressed genes in the four strongest AIR-responding cell types, macrophages, neutrophils, sDCs and mast cells. (C) Venn diagram of ‘not-AIR' transcripts in the four strongest responding cell types. (D) Cell type-specific AIR transcripts are genuinely cell type-specific. AIR transcripts within the ‘any 2 cell types' category were removed from the analysis, but no other constraints were placed. Boxplot outliers are omitted for clarity (See also Fig S9). Mann-Whitney-U test: *p-value < 0.01 between +LPS and +IL-10/+LPS treatments. (E) Box plots showing the changes in gene expression caused by IL-10 on interleukins, chemokines and Tnf family members. Mann-Whitney-U test: *p-value < 0.05 between +LPS and +IL-10/+LPS treatments. (F) Heatmap of genes suppressed by IL-10 in at least three cell types.
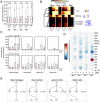
Figure 5. LPS and IL-10 employ different mechanisms in distinct myeloid cells.
(A) CpG percent at the promoters of AIR and not-AIR genes. Promoter is defined as −450 bp +50 bp around the TSS. The numbers of CG dinucleotides were counted within that window and divided by the number of promoters in each cell type. The grey dotted line indicates a CpG island frequency of 0.75. Mann-Whitney-U test, **p-value < 0.01. n.s. not significant. (B) Motif enrichment at the promoters of AIR and not-AIR genes. (C) Changes in expression of Irf family transcripts and Nfkb family transcripts. (D) Boxplots of changes in Irf and Nfkb family transcripts. (E) A putative model for the differing mechanisms stimulated by LPS and IL-10 in the four cell types with both a pro and anti-inflammatory response.
Figure 6. The IL-10/STAT3 target genes are highly divergent in macrophages, neutrophils, sDCs and mast cells.
(A) Venn diagram overlap of genes significantly differentially regulated by IL-10. (B) Heatmap of the 50 genes (60 transcripts) induced by IL-10 in all four cell types. (C) Gene Ontology analysis of IL-10 activated genes in macrophages, neutrophils and sDCs. (D) Selected known IL-10/STAT3 target genes implicated in the AIR. The star indicates a fold-change of at least 1.5 fold in the respective treatment and cell type. Only one Sbno2 transcript is up-regulated whilst at the overall gene level its fold-change is only modestly up-regulated, hence it appears in panel B (transcript analysis).
Similar articles
- Melatonin modulates TLR4-mediated inflammatory genes through MyD88- and TRIF-dependent signaling pathways in lipopolysaccharide-stimulated RAW264.7 cells.
Xia MZ, Liang YL, Wang H, Chen X, Huang YY, Zhang ZH, Chen YH, Zhang C, Zhao M, Xu DX, Song LH. Xia MZ, et al. J Pineal Res. 2012 Nov;53(4):325-34. doi: 10.1111/j.1600-079X.2012.01002.x. Epub 2012 Apr 27. J Pineal Res. 2012. PMID: 22537289 - Understanding and exploiting the endogenous interleukin-10/STAT3-mediated anti-inflammatory response.
Murray PJ. Murray PJ. Curr Opin Pharmacol. 2006 Aug;6(4):379-86. doi: 10.1016/j.coph.2006.01.010. Epub 2006 May 18. Curr Opin Pharmacol. 2006. PMID: 16713356 Review. - The IL-10/STAT3-mediated anti-inflammatory response: recent developments and future challenges.
Hutchins AP, Diez D, Miranda-Saavedra D. Hutchins AP, et al. Brief Funct Genomics. 2013 Nov;12(6):489-98. doi: 10.1093/bfgp/elt028. Epub 2013 Aug 12. Brief Funct Genomics. 2013. PMID: 23943603 Free PMC article. Review.
Cited by
- Models of global gene expression define major domains of cell type and tissue identity.
Hutchins AP, Yang Z, Li Y, He F, Fu X, Wang X, Li D, Liu K, He J, Wang Y, Chen J, Esteban MA, Pei D. Hutchins AP, et al. Nucleic Acids Res. 2017 Mar 17;45(5):2354-2367. doi: 10.1093/nar/gkx054. Nucleic Acids Res. 2017. PMID: 28426095 Free PMC article. - Vitamin C-dependent lysine demethylase 6 (KDM6)-mediated demethylation promotes a chromatin state that supports the endothelial-to-hematopoietic transition.
Zhang T, Huang K, Zhu Y, Wang T, Shan Y, Long B, Li Y, Chen Q, Wang P, Zhao S, Li D, Wu C, Kang B, Gu J, Mai Y, Wang Q, Li J, Zhang Y, Liang Z, Guo L, Wu F, Su S, Wang J, Gao M, Zhong X, Liao B, Chen J, Zhang X, Shu X, Pei D, Nie J, Pan G. Zhang T, et al. J Biol Chem. 2019 Sep 13;294(37):13657-13670. doi: 10.1074/jbc.RA119.009757. Epub 2019 Jul 24. J Biol Chem. 2019. PMID: 31341023 Free PMC article. - MIMIC: an optimization method to identify cell type-specific marker panel for cell sorting.
Zou M, Duren Z, Yuan Q, Li H, Hutchins AP, Wong WH, Wang Y. Zou M, et al. Brief Bioinform. 2021 Nov 5;22(6):bbab235. doi: 10.1093/bib/bbab235. Brief Bioinform. 2021. PMID: 34180954 Free PMC article. - Human and mouse neutrophils share core transcriptional programs in both homeostatic and inflamed contexts.
Hackert NS, Radtke FA, Exner T, Lorenz HM, Müller-Tidow C, Nigrovic PA, Wabnitz G, Grieshaber-Bouyer R. Hackert NS, et al. Nat Commun. 2023 Dec 8;14(1):8133. doi: 10.1038/s41467-023-43573-9. Nat Commun. 2023. PMID: 38065997 Free PMC article. - Artemisia iwayomogi (Dowijigi) inhibits lipopolysaccharide-induced inflammation in RAW264.7 macrophages by suppressing the NF-κB signaling pathway.
Kim SM, Vetrivel P, Kim HH, Ha SE, Saralamma VVG, Kim GS. Kim SM, et al. Exp Ther Med. 2020 Mar;19(3):2161-2170. doi: 10.3892/etm.2020.8472. Epub 2020 Jan 27. Exp Ther Med. 2020. PMID: 32104280 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous