Origins and evolution of viruses of eukaryotes: The ultimate modularity - PubMed (original) (raw)
Review
Origins and evolution of viruses of eukaryotes: The ultimate modularity
Eugene V Koonin et al. Virology. 2015 May.
Abstract
Viruses and other selfish genetic elements are dominant entities in the biosphere, with respect to both physical abundance and genetic diversity. Various selfish elements parasitize on all cellular life forms. The relative abundances of different classes of viruses are dramatically different between prokaryotes and eukaryotes. In prokaryotes, the great majority of viruses possess double-stranded (ds) DNA genomes, with a substantial minority of single-stranded (ss) DNA viruses and only limited presence of RNA viruses. In contrast, in eukaryotes, RNA viruses account for the majority of the virome diversity although ssDNA and dsDNA viruses are common as well. Phylogenomic analysis yields tangible clues for the origins of major classes of eukaryotic viruses and in particular their likely roots in prokaryotes. Specifically, the ancestral genome of positive-strand RNA viruses of eukaryotes might have been assembled de novo from genes derived from prokaryotic retroelements and bacteria although a primordial origin of this class of viruses cannot be ruled out. Different groups of double-stranded RNA viruses derive either from dsRNA bacteriophages or from positive-strand RNA viruses. The eukaryotic ssDNA viruses apparently evolved via a fusion of genes from prokaryotic rolling circle-replicating plasmids and positive-strand RNA viruses. Different families of eukaryotic dsDNA viruses appear to have originated from specific groups of bacteriophages on at least two independent occasions. Polintons, the largest known eukaryotic transposons, predicted to also form virus particles, most likely, were the evolutionary intermediates between bacterial tectiviruses and several groups of eukaryotic dsDNA viruses including the proposed order "Megavirales" that unites diverse families of large and giant viruses. Strikingly, evolution of all classes of eukaryotic viruses appears to have involved fusion between structural and replicative gene modules derived from different sources along with additional acquisitions of diverse genes.
Keywords: Bacteriophages; Evolution of viruses; Functional gene modules; Polintons; Recombination; Transposable elements.
Published by Elsevier Inc.
Figures
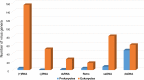
Fig. 1
Representation of different “Baltimore classes” of viruses in prokaryotes and eukaryotes. The bars show the number of genera in the respective classes according to the latest ICTV report (King et al., 2011). Unclassified viruses are disregarded. The numbers for ssDNA viruses also include those for papillomaviruses and polyomaviruses.
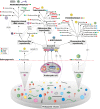
Fig. 2
Origin of the major groups of RNA viruses of eukaryotes. The depicted evolutionary reconstruction is predicated on the symbiogenetic scenario of eukaryogenesis. The host ranges of viral groups are color-coded as shown in the inset. Icons of virion structures are shown for selected groups. Ancestor-descendant relationships that are considered tentative are shown with dotted lines, and particularly weak links are additionally indicated by question marks (see text for details). Key horizontal gene transfer events are shown by gray, curved arrows. Abbreviations: CII FP, Class II fusion protein; CP, capsid protein; CPf, capsid protein of filamentous viruses; JRC, jelly roll capsid (protein); MP, movement protein; RT, reverse transcriptase; S2H, Superfamily 2 helicase; S3H, Superfamily 3 helicase.
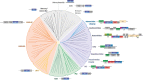
Fig. 3
Evolution of retroelements and reverse-transcribing viruses. Genomic organizations of selected representatives of the major groups of retroelements overlay the phylogenetic tree of the reverse transcriptases. The topology of the tree is from (Gladyshev and Arkhipova, 2011). Abbreviations: DGR, diversity-generating retroelements; X/D/E, maturase, DNA binding, and endonuclease domains, respectively, of the intron-encoded protein; mtd, major tropism determinant; atd, accessory tropism determinant; brt, bacteriophage reverse transcriptase; LINE, long interspersed nucleotide elements; END, endonuclease; ZK, zinc knuckle; gag, group-specific antigen; env, envelope; pol, polymerase; PR, aspartate protease; RT, reverse transcriptase; RH, RNase H; INT, integrase; CHR, chromodomain; MA, matrix protein; CA/Cp, capsid protein; NC, nucleocapsid; 6, 6-kDa protein; vif, vpr, vpu, tat, rev, and nef, regulatory proteins encoded by spliced mRNAs; gp120 and gp41, the 120- (surface) and 41-kDa (transmembrane) glycoproteins; ATF, aphid transmission factor; VAP, virion-associated protein; TT/SR, translation trans-activator/suppressor of RNA interference; TP, terminal protein; P, polymerase; PreS, pre-surface protein (envelope); PX/TA, protein X/transcription activator; trbd, telomerase RNA-binding domain; cc, coiled-coil.
Fig. 4
The conserved RC-Rep proteins of ssDNA viruses and their homologs: key motifs, domain architectures and structures. (A) The catalytic motifs of the nicking endonuclease and superfamily 3 helicase (S3H) domains. Note the absence of the S3H domain in the prokaryotic ssDNA viruses and the inactivation of the endonuclease domain in the dsDNA-containing papillomaviruses and polyomaviruses. (B) Homologous structures of the endonuclease domains. The structures are colored according to the secondary structure elements: α-helixes, blue; β-strands, green. Abbreviations and PDB accession numbers: PCV2, porcine circovirus type 2 (2HW0); TYLCV, tomato yellow leaf curl virus (1L2M); FBNYV, faba bean necrotic yellows virus (2HWT); AAV5, adeno-associated virus type 5 (1M55); SV40, simian virus 40 (1TBD); BPV, bovine papilloma virus (1F08).
Fig. 5
Homologous single jelly-roll structures of the capsid proteins of RNA and DNA viruses of eukaryotes. The structures are colored according to the secondary structure elements: α-helixes, blue; β-strands, green. Depicted structures and their PDB accession numbers: Tymoviridae, turnip yellow mosaic virus (1AUY); Picornaviridae, rhinovirus 16 (1ND2); Satellite virus, satellite tobacco necrosis virus (2BUK); Birnaviridae, infectious bursal disease virus (1WCD); Circoviridae, porcine circovirus type 2 (3R0R); Parvoviridae, adeno-associated virus type 2 (1LP3); Papillomaviridae, human papillomavirus 16 (1DZL); Polyomaviridae, simian virus 40 (1SVA).
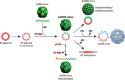
Fig. 6
Evolution of ssDNA viruses of eukaryotes: polyphyletic origin from different plasmids and multiple cases of recombination with ssRNA viruses. Abbreviations: JRC, jelly roll capsid protein; pPolB, protein-primed DNA polymerase of family B; RC-Rep, rolling circle replication protein. Different colors of JRC and RC-Rep denote distinct variants of the respective genes.
Fig. 7
Evolution of large dsDNA viruses of eukaryotes from two distinct groups of bacteriophages. The dotted line with a question mark shows a tenuous evolutionary relationship. The host ranges of the eukaryotic virus groups are color-coded as shown in the inset. The hatched yellow square for the virophages indicates that these viruses parasitize on the giant viruses of the family Mimivirdae which themselves infect amoeba and other protists. For each family of large eukaryotic viruses, a simplified schematic depiction of the virion structure is included.
Similar articles
- Evolution of double-stranded DNA viruses of eukaryotes: from bacteriophages to transposons to giant viruses.
Koonin EV, Krupovic M, Yutin N. Koonin EV, et al. Ann N Y Acad Sci. 2015 Apr;1341(1):10-24. doi: 10.1111/nyas.12728. Epub 2015 Feb 27. Ann N Y Acad Sci. 2015. PMID: 25727355 Free PMC article. Review. - Polintons: a hotbed of eukaryotic virus, transposon and plasmid evolution.
Krupovic M, Koonin EV. Krupovic M, et al. Nat Rev Microbiol. 2015 Feb;13(2):105-15. doi: 10.1038/nrmicro3389. Epub 2014 Dec 22. Nat Rev Microbiol. 2015. PMID: 25534808 Free PMC article. Review. - Self-synthesizing transposons: unexpected key players in the evolution of viruses and defense systems.
Krupovic M, Koonin EV. Krupovic M, et al. Curr Opin Microbiol. 2016 Jun;31:25-33. doi: 10.1016/j.mib.2016.01.006. Epub 2016 Feb 1. Curr Opin Microbiol. 2016. PMID: 26836982 Free PMC article. Review. - The Double-Stranded DNA Virosphere as a Modular Hierarchical Network of Gene Sharing.
Iranzo J, Krupovic M, Koonin EV. Iranzo J, et al. mBio. 2016 Aug 2;7(4):e00978-16. doi: 10.1128/mBio.00978-16. mBio. 2016. PMID: 27486193 Free PMC article. - [The great virus comeback].
Forterre P. Forterre P. Biol Aujourdhui. 2013;207(3):153-68. doi: 10.1051/jbio/2013018. Epub 2013 Dec 13. Biol Aujourdhui. 2013. PMID: 24330969 Review. French.
Cited by
- The immune modules conserved across the tree of life: Towards a definition of ancestral immunity.
Bernheim A, Cury J, Poirier EZ. Bernheim A, et al. PLoS Biol. 2024 Jul 15;22(7):e3002717. doi: 10.1371/journal.pbio.3002717. eCollection 2024 Jul. PLoS Biol. 2024. PMID: 39008452 Free PMC article. - Invasion and Amplification of Endogenous Retroviruses in Dasyuridae Marsupial Genomes.
Harding EF, Mercer LK, Yan GJH, Waters PD, White PA. Harding EF, et al. Mol Biol Evol. 2024 Aug 2;41(8):msae160. doi: 10.1093/molbev/msae160. Mol Biol Evol. 2024. PMID: 39101626 Free PMC article. - Metatranscriptomic Analysis and In Silico Approach Identified Mycoviruses in the Arbuscular Mycorrhizal Fungus Rhizophagus spp.
Neupane A, Feng C, Feng J, Kafle A, Bücking H, Lee Marzano SY. Neupane A, et al. Viruses. 2018 Dec 12;10(12):707. doi: 10.3390/v10120707. Viruses. 2018. PMID: 30545059 Free PMC article. - Genome-wide identification of Reverse Transcriptase domains of recently inserted endogenous plant pararetrovirus (Caulimoviridae).
de Tomás C, Vicient CM. de Tomás C, et al. Front Plant Sci. 2022 Dec 14;13:1011565. doi: 10.3389/fpls.2022.1011565. eCollection 2022. Front Plant Sci. 2022. PMID: 36589050 Free PMC article. - Primordial Capsid and Spooled ssDNA Genome Structures Unravel Ancestral Events of Eukaryotic Viruses.
Munke A, Kimura K, Tomaru Y, Wang H, Yoshida K, Mito S, Hongo Y, Okamoto K. Munke A, et al. mBio. 2022 Aug 30;13(4):e0015622. doi: 10.1128/mbio.00156-22. Epub 2022 Jul 20. mBio. 2022. PMID: 35856561 Free PMC article.
References
- Ackermann H.W., Prangishvili D. Prokaryote viruses studied by electron microscopy. Arch. Virol. 2012;157(10):1843–1849. - PubMed
- Adriaenssens E.M., Edwards R., Nash J.H., Mahadevan P., Seto D., Ackermann H.W., Lavigne R., Kropinski A.M. Integration of genomic and proteomic analyses in the classification of the Siphoviridae family. Virology. 2014 - PubMed
- Agol V.I. Towards the system of viruses. Biosystems. 1974;6(2):113–132. - PubMed
- Ammar el D., Tsai C.W., Whitfield A.E., Redinbaugh M.G., Hogenhout S.A. Cellular and molecular aspects of rhabdovirus interactions with insect and plant hosts. Annu. Rev. Entomol. 2009;54:447–468. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources