Histone H3 mutations--a special role for H3.3 in tumorigenesis? - PubMed (original) (raw)
Review
Histone H3 mutations--a special role for H3.3 in tumorigenesis?
Satish Kallappagoudar et al. Chromosoma. 2015 Jun.
Abstract
Brain tumors are the most common solid tumors in children. Pediatric high-grade glioma (HGG) accounts for ∼8-12 % of these brain tumors and is a devastating disease as 70-90 % of patients die within 2 years of diagnosis. The failure to advance therapy for these children over the last 30 years is largely due to limited knowledge of the molecular basis for these tumors and a lack of disease models. Recently, sequencing of tumor cells revealed that histone H3 is frequently mutated in pediatric HGG, with up to 78 % of diffuse intrinsic pontine gliomas (DIPGs) carrying K27M and 36 % of non-brainstem gliomas carrying either K27M or G34R/V mutations. Although mutations in many chromatin modifiers have been identified in cancer, this was the first demonstration that histone mutations may be drivers of disease. Subsequent studies have identified high-frequency mutation of histone H3 to K36M in chondroblastomas and to G34W/L in giant cell tumors of bone, which are diseases of adolescents and young adults. Interestingly, the G34 mutations, the K36M mutations, and the majority of K27M mutations occur in genes encoding the replacement histone H3.3. Here, we review the peculiar characteristics of histone H3.3 and use this information as a backdrop to highlight current thinking about how the identified mutations may contribute to disease development.
Figures
Fig. 1
Histone H3.3 shows amino acid differences with H3.1 that promote binding to distinct chaperones. a Sequence alignment of human H3.3, H3.2, and H3.1, with sequence differences in H3.3 marked in red. b Cartoon of chromosome depicting regions of H3.3 incorporation into chromatin and the chaperones responsible
Fig. 2
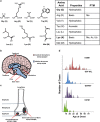
Specific histone H3 mutants arise in distinct regions of the brain or in different skeletal tissues and show variable age of presentation. a The amino acids that substitute glycine at amino acid 34 or lysine at amino acids 27 and 36 of histone H3, their properties, and possible posttranslational modifications. b Cartoon depicting the different anatomical location of brain tumors bearing K27M mutant H3.1 or H3.3 and G34R or G34V mutant H3.3. K27M mutants are predominantly found in midline structures (including the thalamus, pons, and brainstem), whereas the G34 mutant tumors are most commonly located in the cerebral hemispheres (Sturm et al. ; Bjerke et al. 2013). c Cartoon illustrating the distribution of different histone H3 mutants in chondroblastomas and giant cell tumors of bone (Behjati et al. 2013). d Graphical representation of span of age of presentation for histone H3 mutant tumors (Schwartzentruber et al. ; Sturm et al. ; Behjati et al. 2013)
Fig. 3
Codon usage in histone H3.3 and H3.1 genes at the sites of histone mutation. Orange-shaded boxes mark the genes in which mutations are prevalent for the different amino acid substitutions
Fig. 4
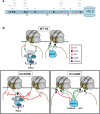
K27M mutants dominantly block PRC2 methyltransferase activity on H3K27, whereas G34R/V mutants block SETD2 methyltransferase function on K36 of the same tail. a Representation of the amino terminal tail of histone H3.3 showing the position of known posttranslational modifications, the site of amino acid substitutions identified in tumors (K27 and G34: red), and the amino acid that differs in the H3.3 tail from H3.1 (Ser 31: orange). b Cartoon depicting the distinct modes of action of K27M mutants and G34R/V mutants in modulating posttranslational modifications on H3 proteins. Note that for the K27M mutant, we depict EZH2 as bound to mutant chromatin, with methyltransferase activity blocked on adjacent sites. Such binding of EZH2 ON chromatin may block the chromatin template from additional chromatin transactions. An alternative possibility is that non-nucleosomal K27M mutant H3 could sequester EZH2 off chromatin, which would leave open the possibility of additional modifications occurring on the mutant chromatin template
Similar articles
- K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas.
Khuong-Quang DA, Buczkowicz P, Rakopoulos P, Liu XY, Fontebasso AM, Bouffet E, Bartels U, Albrecht S, Schwartzentruber J, Letourneau L, Bourgey M, Bourque G, Montpetit A, Bourret G, Lepage P, Fleming A, Lichter P, Kool M, von Deimling A, Sturm D, Korshunov A, Faury D, Jones DT, Majewski J, Pfister SM, Jabado N, Hawkins C. Khuong-Quang DA, et al. Acta Neuropathol. 2012 Sep;124(3):439-47. doi: 10.1007/s00401-012-0998-0. Epub 2012 Jun 3. Acta Neuropathol. 2012. PMID: 22661320 Free PMC article. - Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes.
Castel D, Philippe C, Calmon R, Le Dret L, Truffaux N, Boddaert N, Pagès M, Taylor KR, Saulnier P, Lacroix L, Mackay A, Jones C, Sainte-Rose C, Blauwblomme T, Andreiuolo F, Puget S, Grill J, Varlet P, Debily MA. Castel D, et al. Acta Neuropathol. 2015 Dec;130(6):815-27. doi: 10.1007/s00401-015-1478-0. Epub 2015 Sep 23. Acta Neuropathol. 2015. PMID: 26399631 Free PMC article. - Base-resolution methylomes of gliomas bearing histone H3.3 mutations reveal a G34 mutant-specific signature shared with bone tumors.
Sangatsuda Y, Miura F, Araki H, Mizoguchi M, Hata N, Kuga D, Hatae R, Akagi Y, Amemiya T, Fujioka Y, Arai Y, Yoshida A, Shibata T, Yoshimoto K, Iihara K, Ito T. Sangatsuda Y, et al. Sci Rep. 2020 Sep 30;10(1):16162. doi: 10.1038/s41598-020-73116-x. Sci Rep. 2020. PMID: 32999376 Free PMC article. - Histone H3 Mutations: An Updated View of Their Role in Chromatin Deregulation and Cancer.
Lowe BR, Maxham LA, Hamey JJ, Wilkins MR, Partridge JF. Lowe BR, et al. Cancers (Basel). 2019 May 13;11(5):660. doi: 10.3390/cancers11050660. Cancers (Basel). 2019. PMID: 31086012 Free PMC article. Review. - Histone H3K27M Mutation in Brain Tumors.
El-Hashash AHK. El-Hashash AHK. Adv Exp Med Biol. 2021;1283:43-52. doi: 10.1007/978-981-15-8104-5_3. Adv Exp Med Biol. 2021. PMID: 33155136 Review.
Cited by
- Pediatric glioblastoma cells are sensitive to drugs that inhibit eIF2α dephosphorylation and its phosphomimetic S51D variant.
Eytan K, Versano Z, Oren R, Jacob-Hirsch J, Leitner M, Harmelin A, Rechavi G, Toren A, Paglin S, Yalon M. Eytan K, et al. Front Oncol. 2022 Aug 26;12:959133. doi: 10.3389/fonc.2022.959133. eCollection 2022. Front Oncol. 2022. PMID: 36091130 Free PMC article. - Global activation of oncogenic pathways underlies therapy resistance in diffuse midline glioma.
Georgescu MM, Islam MZ, Li Y, Circu ML, Traylor J, Notarianni CM, Kline CN, Burns DK. Georgescu MM, et al. Acta Neuropathol Commun. 2020 Jul 17;8(1):111. doi: 10.1186/s40478-020-00992-9. Acta Neuropathol Commun. 2020. PMID: 32680567 Free PMC article. - Histone H3G34R mutation causes replication stress, homologous recombination defects and genomic instability in S. pombe.
Yadav RK, Jablonowski CM, Fernandez AG, Lowe BR, Henry RA, Finkelstein D, Barnum KJ, Pidoux AL, Kuo YM, Huang J, O'Connell MJ, Andrews AJ, Onar-Thomas A, Allshire RC, Partridge JF. Yadav RK, et al. Elife. 2017 Jul 18;6:e27406. doi: 10.7554/eLife.27406. Elife. 2017. PMID: 28718400 Free PMC article. - PRC2-independent actions of H3.3K27M in embryonic stem cell differentiation.
Cohen LRZ, Kaffe B, Deri E, Leibson C, Nissim-Rafinia M, Maman M, Harpaz N, Ron G, Shema E, Meshorer E. Cohen LRZ, et al. Nucleic Acids Res. 2023 Feb 28;51(4):1662-1673. doi: 10.1093/nar/gkac800. Nucleic Acids Res. 2023. PMID: 36156096 Free PMC article. - Oncogenic Mechanisms of Histone H3 Mutations.
Weinberg DN, Allis CD, Lu C. Weinberg DN, et al. Cold Spring Harb Perspect Med. 2017 Jan 3;7(1):a026443. doi: 10.1101/cshperspect.a026443. Cold Spring Harb Perspect Med. 2017. PMID: 27864305 Free PMC article. Review.
References
- Adam S, Polo SE, Almouzni G. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell. 2013;155:94–106. - PubMed
- Adam S, Polo SE, Almouzni G. How to restore chromatin structure and function in response to DNA damage—let the chaperones play: delivered on 9 July 2013 at the 38th FEBS Congress in St Petersburg, Russia. FEBS J. 2014;281:2315–2323. - PubMed
- Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. - PubMed
- Bechet D, Gielen GG, Korshunov A, Pfister SM, Rousso C, Faury D, Fiset PO, Benlimane N, Lewis PW, Lu C, David AC, Kieran MW, Ligon KL, Pietsch T, Ellezam B, Albrecht S, Jabado N. Specific detection of methionine 27 mutation in histone 3 variants (H3K27M) in fixed tissue from high-grade astrocytomas. Acta Neuropathol. 2014;128:733–741. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources