Genetic and functional characterization of clonally derived adult human brown adipocytes - PubMed (original) (raw)
doi: 10.1038/nm.3819. Epub 2015 Mar 16.
Ineke H N Luijten 2, Yutaka Hasegawa 1, Haemin Hong 1, Si B Sonne 1, Miae Kim 1, Ruidan Xue 3, Maria Chondronikola 4, Aaron M Cypess 3, Yu-Hua Tseng 3, Jan Nedergaard 5, Labros S Sidossis 4, Shingo Kajimura 1
Affiliations
- PMID: 25774848
- PMCID: PMC4427356
- DOI: 10.1038/nm.3819
Genetic and functional characterization of clonally derived adult human brown adipocytes
Kosaku Shinoda et al. Nat Med. 2015 Apr.
Abstract
Brown adipose tissue (BAT) acts in mammals as a natural defense system against hypothermia, and its activation to a state of increased energy expenditure is believed to protect against the development of obesity. Even though the existence of BAT in adult humans has been widely appreciated, its cellular origin and molecular identity remain elusive largely because of high cellular heterogeneity within various adipose tissue depots. To understand the nature of adult human brown adipocytes at single cell resolution, we isolated clonally derived adipocytes from stromal vascular fractions of adult human BAT from two individuals and globally analyzed their molecular signatures. We used RNA sequencing followed by unbiased genome-wide expression analyses and found that a population of uncoupling protein 1 (UCP1)-positive human adipocytes possessed molecular signatures resembling those of a recruitable form of thermogenic adipocytes (that is, beige adipocytes). In addition, we identified molecular markers that were highly enriched in UCP1-positive human adipocytes, a set that included potassium channel K3 (KCNK3) and mitochondrial tumor suppressor 1 (MTUS1). Further, we functionally characterized these two markers using a loss-of-function approach and found that KCNK3 and MTUS1 were required for beige adipocyte differentiation and thermogenic function. The results of this study present new opportunities for human BAT research, such as facilitating cell-based disease modeling and unbiased screens for thermogenic regulators.
Figures
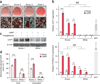
Figure 1
Isolation of clonal brown adipocytes from adult human BAT. (a) Representative Oil-Red-O staining of differentiated brown adipocyte cultures 1–3 and white adipocyte culture 1 at low magnification (top) and at high magnification (bottom). n = 3 for all groups. Scale bars, 50 µm. (b) Expression of UCP1 (top) and PPARGC1A (bottom) in the differentiated clonal brown adipocyte cultures 1–3 and white adipocyte cultures 1–3 treated with forskolin (cAMP) or vehicle (basal). BAT, biopsied human BAT from the supraclavicular regions (positive control). mRNA expression relative to expression of housekeeping gene TBP. n = 3 for all groups. §§§P < 0.001, brown versus white adipocyte lines; *P < 0.05, **P < 0.01, ***P < 0.001, brown or white (as indicated) versus basal by one-sided Welch’s _t_-test. The error bars for UCP1 in white adipocytes are 0.001, 0.002 and 0.001 in cultures 1, 2 and 3, respectively. (c) Western blot of UCP1 in differentiated brown adipocyte culture 1 and white adipocyte culture 1 treated with forskolin (cAMP) or rosiglitazone. β-actin, loading control. Data are representative of two experiments. (d) Total and uncoupled cellular respiration in differentiated brown adipocyte culture 2 and white adipocyte culture 1 treated with forskolin (cAMP) or vehicle (basal). OCR, oxygen consumption rate. n = 8 for all groups. ***P < 0.001 by one-sided Student’s _t_-test. NS, not significant. The variance was similar between basal and cAMP groups (P = 0.709). Data are expressed as means ± s.e.m. for all bar graphs.
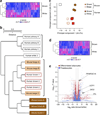
Figure 2
Genome-wide gene expression analyses indicate a close relationship between human brown adipocytes and mouse beige adipocytes. (a) Expression profile and hierarchical clustering of the differentially expressed genes between differentiated clonal human brown adipocyte cultures 1–3 and differentiated clonal white adipocyte cultures 1–3 by two-fold or more. n = 3 for each cell type. The color scale shows _z_-scored FPKM (fragments per kilobase of exon per million fragments mapped) representing the mRNA level of each gene in a blue (low expression)-white-red (high expression) scheme. (b) Hierarchical clustering of human and mouse adipocytes as visualized by TreeGraph. The horizontal distance represents similarities among each cluster. (c) Principal component (PC) analysis of the transcriptome from human and mouse differentiated adipocytes. PC analysis was done using the same gene expression data set used in b, that is, the RNA-seq data set obtained from differentiated clonal human brown and white adipocyte cultures, and the microarray data set (
GSE39562
) from differentiated clonal mouse classical brown and beige adipocytes. Numbers in parentheses represent the proportion of data variance explained by each PC. (d) Expression profiles and hierarchical clustering of the differentially expressed genes between undifferentiated clonal human brown preadipocyte cell lines 1–3 and white preadipocyte cell lines 1–3 by two-fold or more. n = 3 for each cell type. The color scale is the same as in a. (e) Volcano plot of transcriptomes in the clonal differentiated human brown and white adipocyte cultures (red) and in the clonal undifferentiated human brown and white preadipocyte lines (blue). n = 3 for each cell type. The log-fold change between brown versus white is shown on the _x_-axis. The _y_-axis represents the −log10of the P values by delta method–based test. Previously defined BAT-enriched markers are shown.
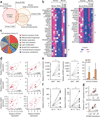
Figure 3
Identification of human brown adipocyte markers. (a) Venn diagram of the overlapping genes enriched in human brown adipocytes, mouse classical brown adipocytes, and mouse beige adipocytes versus white adipocytes of the respective species by two-fold or more. All cells were differentiated clonal adipocytes in culture. P < 0.05 by delta method–based test. (b) Expression profiles of select genes enriched in each group. The color scale shows _z_-scored FPKM representing the mRNA level of each gene in blue (low expression)-white-red (high expression) scheme. (c) GO analysis of the gene set in Group A (GO FAT category). The area of each pie slice represents the number of genes that belong to the indicated GO terms. (d) Correlation between MTUS1 and KCNK3 mRNA expression on the _x_-axis and mRNA expression of previously defined marker genes PPARGC1A, PRDM16 and CIDEA on the _y_-axis. mRNA expression relative to the housekeeping gene TBP. n = 23 for each panel. P < 0.01 by _z_-test. (e) Gene expression of MTUS1 and KCNK3 in UCP1-positive adipose tissues (BAT) and UCP1-negative adipose tissues (WAT) from the neck region of the same individuals (eight pairs). *P = 0.017 and 0.044, respectively, by Wilcoxon signed-ranked test. The right bar graph shows the expression data without normalization to each individual. Data are expressed as means ± s.e.m. n = 8 for each group, *P < 0.05 by one-sided Welch’s _t_-test. (f) mRNA expression of MTUS1 and KCNK3 in the supraclavicular BAT isolated from six subjects under thermoneutral conditions (30 °C) and prolonged cold exposure (19 °C). Expression relative to TBP. *P = 0.035 and 0.023, respectively, by Wilcoxon signed-ranked test. (g) Correlation analysis between MTUS1 variant 3 or KCNK3 and UCP1 under prolonged cold exposure. n = 13. P < 0.01 by _z_-test.
Figure 4
Mtus1 and Kcnk3 are required for beige adipocyte differentiation and thermogenic function. (a) Expression of Mtus1, Kcnk3, and Fzd8 in mouse inguinal WAT-derived adipocytes transfected with siRNAs targeting the indicated genes or non-targeting control (Ctrl). n = 4 for all groups. ***P < 0.001 for indicated siRNA versus control by one-sided Welch’s _t_-test. (b) Ucp1, Mtus1 and Kcnk3 protein expression from cells under the same culture condition as in a. β-actin, loading control. (c) Expression of Ucp1, Cidea, Cox7a1, and Adipoq (encoding adiponectin) from cells under the same culture condition as in a. n = 4 for all groups. **P < 0.01, ***P < 0.001 indicated siRNA versus control by one-sided Student’s _t_-test. NS, not significant. (d) Total and uncoupled cellular respiration. n = 8 for all groups. **P < 0.01, ***P < 0.001 by one-sided Welch’s _t_-test. Data are expressed as means ± s.e.m. for all bar graphs. Data are representative of three independent experiments.
Similar articles
- Functional thermogenic beige adipogenesis is inducible in human neck fat.
Lee P, Werner CD, Kebebew E, Celi FS. Lee P, et al. Int J Obes (Lond). 2014 Feb;38(2):170-6. doi: 10.1038/ijo.2013.82. Epub 2013 May 21. Int J Obes (Lond). 2014. PMID: 23736373 Free PMC article. - The K+ channel TASK1 modulates β-adrenergic response in brown adipose tissue through the mineralocorticoid receptor pathway.
Pisani DF, Beranger GE, Corinus A, Giroud M, Ghandour RA, Altirriba J, Chambard JC, Mazure NM, Bendahhou S, Duranton C, Michiels JF, Frontini A, Rohner-Jeanrenaud F, Cinti S, Christian M, Barhanin J, Amri EZ. Pisani DF, et al. FASEB J. 2016 Feb;30(2):909-22. doi: 10.1096/fj.15-277475. Epub 2015 Nov 2. FASEB J. 2016. PMID: 26527067 - Adaptive thermogenesis in brown adipose tissue involves activation of pannexin-1 channels.
Senthivinayagam S, Serbulea V, Upchurch CM, Polanowska-Grabowska R, Mendu SK, Sahu S, Jayaguru P, Aylor KW, Chordia MD, Steinberg L, Oberholtzer N, Uchiyama S, Inada N, Lorenz UM, Harris TE, Keller SR, Meher AK, Kadl A, Desai BN, Kundu BK, Leitinger N. Senthivinayagam S, et al. Mol Metab. 2021 Feb;44:101130. doi: 10.1016/j.molmet.2020.101130. Epub 2020 Nov 25. Mol Metab. 2021. PMID: 33248294 Free PMC article. - UCP1 Dependent and Independent Thermogenesis in Brown and Beige Adipocytes.
Ikeda K, Yamada T. Ikeda K, et al. Front Endocrinol (Lausanne). 2020 Jul 28;11:498. doi: 10.3389/fendo.2020.00498. eCollection 2020. Front Endocrinol (Lausanne). 2020. PMID: 32849287 Free PMC article. Review. - Adaptive thermogenesis in adipocytes: is beige the new brown?
Wu J, Cohen P, Spiegelman BM. Wu J, et al. Genes Dev. 2013 Feb 1;27(3):234-50. doi: 10.1101/gad.211649.112. Genes Dev. 2013. PMID: 23388824 Free PMC article. Review.
Cited by
- Sortilin-mediated translocation of mitochondrial ACSL1 impairs adipocyte thermogenesis and energy expenditure in male mice.
Yang M, Ge J, Liu YL, Wang HY, Wang ZH, Li DP, He R, Xie YY, Deng HY, Peng XM, Wang WS, Liu JD, Zhu ZZ, Yu XF, Maretich P, Kajimura S, Pan RP, Chen Y. Yang M, et al. Nat Commun. 2024 Sep 5;15(1):7746. doi: 10.1038/s41467-024-52218-4. Nat Commun. 2024. PMID: 39232011 Free PMC article. - Transcriptome analysis reveals brown adipogenic reprogramming in chemical compound-induced brown adipocytes converted from human dermal fibroblasts.
Takeda Y, Yoshikawa T, Dai P. Takeda Y, et al. Sci Rep. 2021 Mar 3;11(1):5061. doi: 10.1038/s41598-021-84611-0. Sci Rep. 2021. PMID: 33658606 Free PMC article. - Mitochondrial Lipid Signaling and Adaptive Thermogenesis.
Von Bank H, Hurtado-Thiele M, Oshimura N, Simcox J. Von Bank H, et al. Metabolites. 2021 Feb 22;11(2):124. doi: 10.3390/metabo11020124. Metabolites. 2021. PMID: 33671745 Free PMC article. Review. - Beige Adipocyte Maintenance Is Regulated by Autophagy-Induced Mitochondrial Clearance.
Altshuler-Keylin S, Shinoda K, Hasegawa Y, Ikeda K, Hong H, Kang Q, Yang Y, Perera RM, Debnath J, Kajimura S. Altshuler-Keylin S, et al. Cell Metab. 2016 Sep 13;24(3):402-419. doi: 10.1016/j.cmet.2016.08.002. Epub 2016 Aug 25. Cell Metab. 2016. PMID: 27568548 Free PMC article. - Adenosine/A2B Receptor Signaling Ameliorates the Effects of Aging and Counteracts Obesity.
Gnad T, Navarro G, Lahesmaa M, Reverte-Salisa L, Copperi F, Cordomi A, Naumann J, Hochhäuser A, Haufs-Brusberg S, Wenzel D, Suhr F, Jespersen NZ, Scheele C, Tsvilovskyy V, Brinkmann C, Rittweger J, Dani C, Kranz M, Deuther-Conrad W, Eltzschig HK, Niemi T, Taittonen M, Brust P, Nuutila P, Pardo L, Fleischmann BK, Blüher M, Franco R, Bloch W, Virtanen KA, Pfeifer A. Gnad T, et al. Cell Metab. 2020 Jul 7;32(1):56-70.e7. doi: 10.1016/j.cmet.2020.06.006. Epub 2020 Jun 25. Cell Metab. 2020. PMID: 32589947 Free PMC article.
References
- Ouellet V, et al. Outdoor temperature age, sex, body mass index, and diabetic status determine the prevalence, mass, and glucose-uptake activity of 18F-FDG-detected BAT in humans. J. Clin. Endocrinol. Metab. 2011;96:192–199. - PubMed
- van Marken Lichtenbelt WD, et al. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 2009;360:1500–1508. - PubMed
- Yoneshiro T, et al. Age-related decrease in cold-activated brown adipose tissue and accumulation of body fat in healthy humans. Obesity (Silver Spring) 2011;19:1755–1760. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK097441/DK/NIDDK NIH HHS/United States
- DK087853/DK/NIDDK NIH HHS/United States
- K99 DK087853/DK/NIDDK NIH HHS/United States
- P50-GM60338/GM/NIGMS NIH HHS/United States
- DK097441/DK/NIDDK NIH HHS/United States
- P50 GM060338/GM/NIGMS NIH HHS/United States
- R00 DK087853/DK/NIDDK NIH HHS/United States
- P30 DK063720/DK/NIDDK NIH HHS/United States
- DK63720/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials