The fine-scale genetic structure of the British population - PubMed (original) (raw)
. 2015 Mar 19;519(7543):309-314.
doi: 10.1038/nature14230.
Bruce Winney # 3, Garrett Hellenthal # 4, Dan Davison 5, Abdelhamid Boumertit 3, Tammy Day 3, Katarzyna Hutnik 3, Ellen C Royrvik 3, Barry Cunliffe 6; Wellcome Trust Case Control Consortium 2; International Multiple Sclerosis Genetics Consortium; Daniel J Lawson 7, Daniel Falush 8, Colin Freeman 9, Matti Pirinen 10, Simon Myers 11, Mark Robinson 12, Peter Donnelly 9 11, Walter Bodmer 3
Affiliations
- PMID: 25788095
- PMCID: PMC4632200
- DOI: 10.1038/nature14230
The fine-scale genetic structure of the British population
Stephen Leslie et al. Nature. 2015.
Abstract
Fine-scale genetic variation between human populations is interesting as a signature of historical demographic events and because of its potential for confounding disease studies. We use haplotype-based statistical methods to analyse genome-wide single nucleotide polymorphism (SNP) data from a carefully chosen geographically diverse sample of 2,039 individuals from the United Kingdom. This reveals a rich and detailed pattern of genetic differentiation with remarkable concordance between genetic clusters and geography. The regional genetic differentiation and differing patterns of shared ancestry with 6,209 individuals from across Europe carry clear signals of historical demographic events. We estimate the genetic contribution to southeastern England from Anglo-Saxon migrations to be under half, and identify the regions not carrying genetic material from these migrations. We suggest significant pre-Roman but post-Mesolithic movement into southeastern England from continental Europe, and show that in non-Saxon parts of the United Kingdom, there exist genetically differentiated subgroups rather than a general 'Celtic' population.
Figures
**Extended Data Figure 1.. The effect of setting a threshold on the confidence of cluster assignment for the genetic clusters in the UK inferred by the fineSTRUCTURE analysis
The UK map depicts the clustering of the 2,039 UK individuals into 17 clusters on the basis of genetics alone. See Figure 1 for further details. Here a threshold is set on the measurement of confidence used for assigning individuals to clusters (see Methods). This measure is defined on the interval [0, 1], where the value 1 is interpreted as meaning complete certainty of cluster assignment and 0 as being complete lack of certainty. The plot illustrates the effect of setting a threshold of 0.7 so that a UK individual is only assigned to a cluster if the measure of assignment for that individual is greater than 0.7. All of the samples that have small, faded symbols are assigned to their clusters with confidence greater than 0.7. Those samples for which the assignment is less confident (i.e. the measure is less than or equal to 0.7) are plotted with large, bold symbols. The table shows the number of individuals with confidence measure above and below the 0.7 threshold together with the total for each UK cluster. The slight discrepancy between the totals in this figure and Extended Data Fig. 3.16 is due to differences in the method for assigning individuals to clusters (see Methods). The threshold of 0.7 was chosen for illustrative purposes only. Similar patterns relate to other thresholds.
**Extended Data Figure 2.. Convergence of the algorithm implemented in fineSTRUCTURE
The fineSTRUCTURE clustering algorithm was run twice on the UK samples (a) and twice on the European samples (b) to assess convergence. The displayed heatmap depicts the proportion of sampled MCMC iterations for which each pair of UK individuals is assigned to the same cluster. The values above and below the diagonal represent two different runs of fineSTRUCTURE. Individuals are ordered along each axis according to the inferred tree from the fineSTRUCTURE run above the diagonal, with tick-marks on the axes at the middle of each cluster. Comparison between runs is made by comparing the plot above the diagonal (run two) with that below the diagonal (run one). The high degree of symmetry in the plot confirms the similarity between the runs and hence that each MCMC run has converged to very similar clusters.
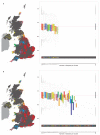
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figures 3.1 - 3.24.. Genetic clusters in the UK inferred by the fineSTRUCTURE analysis at all levels of the hierarchical clustering
Each of the plots 3.1 – 3.23 shows exactly the same information, but for different numbers of clusters, from 2 to 24 in order, determined by the hierarchical clustering analysis. At the level of 24 clusters every cluster has at least 10 members. This is not the case for finer levels of clustering and for brevity these levels are omitted. The final figure, 3.24 shows the final clustering by fineSTRUCTURE, with 53 clusters. a, The UK map depicts the clustering of the 2,039 UK individuals into clusters on the basis of genetics alone. Each symbol corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each genetic cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster. No relationship between clusters is implied by the colours/symbols. In addition there is a table at each level that displays the number of samples in each of the inferred clusters. b, A tree depicting the order of the merging of the clusters in the hierarchical clustering. The cluster symbols are the same as shown in a. The lengths of the branches relate to changes in the likelihood of the statistical model underlying fineSTRUCTURE. They do not relate directly to time or other measures of genetic distance so caution is needed in their interpretation. Some additional length is added to the tips of the tree for clarity. c, The UK samples plotted against the first two principal components as determined in the genome-wide principal components analysis. For comparison, each individual is depicted by the same symbol as in the fineSTRUCTURE analysis depicted in a. The ellipses are drawn as in a.
**Extended Data Figure 4.. Application of standard methods for detecting population structure to the UK data
a, Genome-wide principal component analysis of the UK samples. The UK samples plotted against all pairs of principal component axes, for the first five axes, as determined in the genome-wide principal components analysis. Each individual is depicted by a symbol representing the district from which it was collected. The labels of the sample collection districts are interpreted as follows: CUM = Cumbria; LIN = Lincolnshire; NEA = North East England; OXF = Oxfordshire; YOR = Yorkshire; CHE = Cheshire; NTH = Northamptonshire; NOT = Nottinghamshire; DOR = Dorset; SUS = Sussex; NOR = Norfolk; WOR = Worcestershire; DEV = Devon; SPE = South Pembrokeshire; COR = Cornwall; NWA = North Wales; ARG = Argyle and Bute; NPE = North Pembrokeshire; BAN = Banff and Buchan; NIR = Northern Ireland; ORK = Orkney; SUF = Suffolk; LEI = Leicestershire; FOD = Forest of Dean; HER = Herefordshire; HAM = Hampshire; DER = Derbyshire; LAN = Lancashire; KEN = Kent; GLO = Gloucestershire. b, Clustering the UK samples using the program ADMIXTURE. ADMIXTURE was applied in three scenarios, corresponding to different preset values for K, the number of clusters into which the UK samples are divided. Here K = 2, 3 and 17 (see Methods). A map is shown for each value of K. Each symbol on the map corresponds to one of the sampled individuals and is plotted at the centroid of their grandparents’ birthplace. Each cluster is represented by a unique combination of colour and plotting symbol, with individuals depicted with the symbol of the cluster to which they are assigned. The ellipses centred on each cluster give a sense of the extent of the cluster by showing the 90% probability region of the two-dimensional t-distribution (5 degrees of freedom) which best fits the locations of the individuals in the cluster.
**Extended Data Figure 5.. Potential recent shared ancestry in the genetic clusters in the UK inferred by the fineSTRUCTURE analysis
a, The UK map to the left depicts the clustering of the 2,039 UK individuals into 17 clusters on the basis of genetics alone. See Fig. 1 for further details. Pairwise identity by descent (IBD) within clusters and across the whole UK sample for all of the 2,039 UK individuals is shown to the right. For each of the inferred UK clusters a box and whisker plot shows the distribution of the pairwise IBD statistic (see Methods). Each box is filled by the colour of the cluster to which it relates, and the outlier points have the same shape as the cluster to which they relate. For comparison the distribution of the pairwise IBD statistic across the whole UK sample is shown on the far right, with the box coloured grey. The light grey horizontal lines indicate the upper and lower quartiles of the IBD statistic’s distribution for the whole UK sample. Along the x-axis the number of samples in the associated cluster is shown. The y-axis gives the value of the pairwise IBD statistic. Note that only clusters of size 4 or less depart substantially from the average relatedness. b, The same information as a but with 53 clusters of UK individuals.
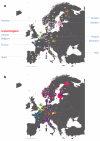
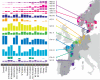
**Extended Data Figure 6.. Population Structure in the European Samples
a, Number of samples derived from each European sampling region. The 6,209 European samples used for the analyses were sampled from ten countries and various locations within each country. Each sample has a specific sampling location (often a city, but in some cases only a whole country). The numbers shown give the number of samples derived from a particular location. Some numbers are depicted out of position for clarity. In these cases a yellow line leads from the number to the actual location. Where the sample locations are well-localized (e.g. the city of sampling is known) the box surrounding the number is white. When only information about the country of sampling is known the box is coloured yellow. The numbers are overlain on a faded version of the pie charts from Extended Data Fig. 6b for easy reference. b, European ancestry profiles of the UK clusters. The 6,209 European samples divided into 51 genetic groups (represented by colours and labelled with a subset of the numbers between 1 and 145) using our fineSTRUCTURE analyses. For clarity the colour space has been skewed to emphasize the differences between groups 1 to 18 as these groups are the major contributors to the ancestry profiles of the UK clusters. Each sample has a specific sampling location (often a city, but in some cases only a country, see Extended Data Fig 6a). The pie charts are located at these sampling locations, and depict the proportion of the samples from that location assigned to each of the 51 genetic groups. Each genetic group also has a label number, which is displayed for the larger sectors of each of the pie charts. The area of the pie chart is proportional to the number of samples from that location. Pie charts with black borders correspond to well-localized samples. In contrast, for samples where only the country of sampling is known, they are combined in a single pie chart for the country, which is shown with white borders. Some pie charts are depicted out of position for clarity; in these cases a yellow line leads from the chart to the actual location.
**Extended Data Figure 7.. European ancestry profiles of the UK clusters
a, The map of the UK shown relates to the map with 17 UK clusters shown in Fig. 1. Ellipses indicate the extent of the UK clusters as in Fig. 1. The pie charts represent the ancestry profile of the UK clusters from Fig. 1. Each pie chart is plotted at the centroid of the corresponding cluster, although some pie charts have been moved for clarity; in the cases where the relocation is substantial a red line leads from the pie chart to the centroid. The sectors of the pie charts are coloured with the colours of the European genetic groups (for the larger sectors the number of the European group is also given). They indicate the ancestry profiles of each UK cluster, namely the proportion of the cluster ancestry that is best represented by each of the European groups. The magnitude of the angle of a sector is proportional to the contribution of that European group to the ancestry profile of the associated UK cluster. The symbols in the grey bar to the left of the map represent the UK clusters as in Fig. 1. The bar chart in the left part of the plot depicts the same ancestry profiles of the UK clusters in a different way. Each row represents a UK cluster (arranged roughly north to south) with the symbols for the clusters from Fig. 1 indicated at each end of the row. Each column represents a European group, with group numbers listed with a three letter prefix that, for clarity, relates to the country or countries where the cluster is most represented. The colour of each bar also indicates the European group to which the bar relates. Confidence intervals (95%) obtained from 1,000 bootstraps of the ancestry profile analysis (see Methods) are indicated on each bar. b, Renormalized ancestry profiles of the UK clusters illustrating possible early European contributions to the UK population. A representation of the relative contributions to the UK clusters from the three European groups (GER6-W. Germany, BEL11- Belgium, and FRA14-NW France) hypothesized to be the major contributors to the earliest migrations into the UK after the last ice age from which DNA survives to the present in substantial proportions (see Supplementary Note). Interpretation of the map, pie charts and bar chart is as for a. In this case, however, the proportions were renormalized to sum to 1 for the contributions from GER6, BEL11 and FRA14.
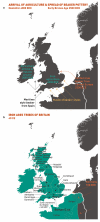
**Extended Data Figure 8.. More major events in the peopling of the British Isles
See Supplementary Note for further details. a, The arrival of agriculture and subsequent migrations from 4000 – 2500 BCE. b, The major Iron Age tribes of Britain around the year 40.
**Extended Data Figure 9.. Application of GLOBETROTTER to infer simulation of ancestry 40 generations ago between groups from Northern Germany (GER 3, 2 5%) and Italy (ITA 36, 75%)
Twenty-five admixed individuals were simulated, and the individuals used to construct these simulated individuals were then removed from the list of potential donors (see Methods). Left barplot, and map: The barplot shows the true population and proportion contributed for each of the two admixing groups. The map shows, for each of the European sampling locations, the true proportion of individuals sampled from that location assigned to each of the admixing groups, coloured according to the barplot. Central three plots: example curves constructed by GLOBETROTTER to infer admixture times, and infer details of admixing groups (see Methods and Supplementary Note). For each pair of populations A and B (A can be the same as or different from B) the points show the empirical probability, relative to under independence, as a function of genetic distance x, that two positions separated by distance x correspond to ancestry donated by population A, and by population B, respectively. The green line shows GLOBETROTTER fitted exponential decay curves for the underlying (i.e. expected) value of this relative probability estimate. Under a model of a single admixture event occurring g generations ago, this probability decays at a rate g according to theory, providing an estimate of the admixture time (and 95% CI) shown overlaying curves ITA 36 v GER 3 and ITA 36 v ITA 36. If ancestries A and B associate with the
same
admixing group - e.g. whenever A=B - the fitted curve will have negative slope, as seen for the GER 3 v GER 3 plot. If a positive slope is seen, as for the ITA 36 v GER 3 plot, this implies these populations contribute to the two
different
respective admixing groups. Right barplot, and map: GLOBETROTTER produces an inference of the genetic composition of (haplotypes carried by) the two admixing groups, as a mixture of (haplotypes carried by) populations actually sampled. This mixture inference jointly uses curves for pairs of sampled populations, and the overall haplotypic makeup of different sampled populations, including the admixed group. The barpot shows the inferred mixture representation (dominated in each case by the true admixing groups) and estimated admixture proportion (24%, close to the truth of 25%), with more red/blue populations respectively giving a larger contribution. The map shows populations inferred as contributing to the first (pink/red shades) or second (blue shades) admixing group, respectively, as for the left map, with populations coloured according to the barplot. This shows populations falsely inferred as contributing material to the admixing groups were still sampled, mainly, from locations close to those of the true admixing groups. We caution that in this setting of admixture between genetically similar European groups, estimation of admixture fraction is very uncertain (see Methods) (e.g. contributing populations are often impossible to definitively assign to a “side” of the event). For further details of the analysis, e.g. tests for admixture presence in this simulation, see Methods and Supplementary Note.
**Extended Data Figure 10.. Application of GLOBETROTTER to infer details of admixture in the UK clusters
a, Inferring admixture in a population of 1,044 UK individuals from central and southern England. Left hand plot: the bold red squares show mean grandparental birthplace for each individual in this population. Central three plots: example curves constructed by GLOBETROTTER to infer admixture times, and infer details of admixing groups (see Methods and Supplementary Note). For each pair of populations A and B (A can be the same as or different from B) the points show the empirical probability, relative to under independence, as a function of genetic distance x, that two positions separated by distance x correspond to ancestry donated by population A, and by population B, respectively. The green line shows GLOBETROTTER fitted exponential decay curves for the underlying (i.e. expected) value of this relative probability estimate. Under a model of a single admixture event occurring g generations ago, this probability decays at a rate g according to theory, providing an estimate of the admixture time (and 95% CI) shown overlaying curves SFS 31 v GER 3 and SFS 31 v SFS 31. If ancestries A and B associate with the
same
admixing group - e.g. whenever A=B - the fitted curve will have negative slope, as seen for the GER 3 v GER 3 plot. If a positive slope is seen, as for the SFS 31 v GER 3 plot, this implies these populations contribute to the two
different
respective admixing groups. Right barplot, and map: GLOBETROTTER inference shows one possibility for the genetic composition of (haplotypes carried by) the two unsampled historical admixing groups, as a mixture of (haplotypes carried by) populations actually sampled. This mixture inference jointly uses curves for pairs of sampled populations, and the overall haplotypic makeup of different sampled populations, including the admixed group. The barpot shows the inferred mixture representation (with largest contributions in each case by GER3/DEN18, sampled most frequently from northern Germany and Denmark, and SFS31/ITA52, sampled mainly from southern France and Spain and northern Italy) and estimated admixture proportion (34%), with more intense red/blue populations respectively implying a larger contribution. The map shows populations inferred as contributing to the first (pink/red shades) or second (blue shades) admixing group respectively, with populations coloured according to the barplot. We caution that in this setting of admixture between genetically similar European groups, estimation of admixture fraction is very uncertain (see Methods and Supplementary Note) (e.g. contributing populations are often impossible to definitively assign to a “side” of the event), so that other closely related scenarios, e.g. a somewhat lower admixture fraction from a more completely “GER 3” like group than that inferred, are likely consistent with the GLOBETROTTER results seen. b, Inferring admixture in a population of 51 UK individuals from Orkney. Left hand plot: the bold purple squares show mean grandparental birthplace for each individual in this population. Central three plots: example curves constructed by GLOBETROTTER to infer admixture times, and infer details of admixing groups (see Methods and Supplementary Note). For each pair of populations A and B (A can be the same as or different from B) the points show the empirical probability, relative to under independence, as a function of genetic distance x, that two positions separated by distance x correspond to ancestry donated by population A, and by population B, respectively. The green line shows GLOBETROTTER fitted exponential decay curves for the underlying (i.e. expected) value of this relative probability estimate. Under a model of a single admixture event occurring g generations ago, this probability decays at a rate g according to theory, providing an estimate of the admixture time (and 95% CI) shown overlaying curves NOR 90 v FRA 12 and NOR 90 v NOR 90. If ancestries A and B associate with the
same
admixing group - e.g. whenever A=B - the fitted curve will have negative slope, as seen for the NOR 90 v NOR 90 plot. If a positive slope is seen, as for the NOR 90 v FRA 12 plot, this implies these populations contribute to the two
different
respective admixing groups. Right barplot, and map: GLOBETROTTER inference shows one possibility for the genetic composition of (haplotypes carried by) the two unsampled historical admixing groups, as a mixture of (haplotypes carried by) populations actually sampled. This mixture inference jointly uses curves for pairs of sampled populations, and the overall haplotypic makeup of different sampled populations, including the admixed group. The barpot shows the inferred mixture representation (with largest contribution in each case by GER3/NOR90, sampled most frequently from northern Germany and Norway, and FRA12/FRAC14, both sampled mainly from France) and estimated admixture proportion (42%), with more intense red/blue populations respectively implying a larger contribution. The map shows populations inferred as contributing to the first (pink/red shades) or second (blue shades) admixing group respectively, with populations coloured according to the barplot. We caution that in this setting of admixture between genetically similar European groups, estimation of admixture fraction is very uncertain (see Methods and Supplementary Note) (e.g. contributing populations are often impossible to definitively assign to a “side” of the event). In particular, inspection of curves involving GER 3 does not yield a clear “side” of the event for this population, unlike the NOR 90 v FRA 12 case that implies French-like and Norwegian-like haplotype presence must occur mainly in distinct admixing groups. Therefore the GER 3 component might in fact capture haplotypes for either (or both) the French-like or Norwegian-like admixing groups, and the inferred scenario shows only one possibility.
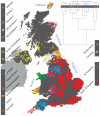
Figure 1. Clustering of the 2,039 UK individuals into 17 clusters based only on genetic data
For each individual, the coloured symbol representing the genetic cluster to which the individual is assigned is plotted at the centroid of their grandparents’ birthplace. Cluster names are in side-bars and ellipses give an informal sense of the range of each cluster (see Methods). No relationship between clusters is implied by the colours/symbols. The tree (top right) depicts the order of the hierarchical merging of clusters (see Methods for the interpretation of branch lengths).
Figure 2. European ancestry profiles for the 17 UK clusters
Each row represents a European group (labels at right). Each column represents a UK cluster. Coloured bars have heights representing the proportion of the UK cluster’s ancestry best represented by that of the European group labelled with that colour. The map shows the location (when known at regional level) of the samples assigned to each European group (some sample locations are jittered and/or moved for clarity, see Methods). Lines join group labels to the centroid of the group, or collection of groups (Norway, Sweden, with individual group centroids marked by group number).
Figure 3. Major events in the peopling of the British Isles
See Supplementary Note for further details. a, The routes taken by the first settlers after the last ice age. b, Britain during the period of Roman rule. c, The regions of ancient British, Irish and Saxon control. d, The migrations of Norse and Danish Vikings. The main regions of Norse Viking (light brown) and Danish Viking (light blue) settlement are shown.
Comment in
- Population genetics. The peopling of Britain.
Skipper M. Skipper M. Nat Rev Genet. 2015 May;16(5):256-7. doi: 10.1038/nrg3938. Epub 2015 Mar 31. Nat Rev Genet. 2015. PMID: 25824870 No abstract available.
Similar articles
- Iron Age and Anglo-Saxon genomes from East England reveal British migration history.
Schiffels S, Haak W, Paajanen P, Llamas B, Popescu E, Loe L, Clarke R, Lyons A, Mortimer R, Sayer D, Tyler-Smith C, Cooper A, Durbin R. Schiffels S, et al. Nat Commun. 2016 Jan 19;7:10408. doi: 10.1038/ncomms10408. Nat Commun. 2016. PMID: 26783965 Free PMC article. - Insular Celtic population structure and genomic footprints of migration.
Byrne RP, Martiniano R, Cassidy LM, Carrigan M, Hellenthal G, Hardiman O, Bradley DG, McLaughlin RL. Byrne RP, et al. PLoS Genet. 2018 Jan 25;14(1):e1007152. doi: 10.1371/journal.pgen.1007152. eCollection 2018 Jan. PLoS Genet. 2018. PMID: 29370172 Free PMC article. - Genomic signals of migration and continuity in Britain before the Anglo-Saxons.
Martiniano R, Caffell A, Holst M, Hunter-Mann K, Montgomery J, Müldner G, McLaughlin RL, Teasdale MD, van Rheenen W, Veldink JH, van den Berg LH, Hardiman O, Carroll M, Roskams S, Oxley J, Morgan C, Thomas MG, Barnes I, McDonnell C, Collins MJ, Bradley DG. Martiniano R, et al. Nat Commun. 2016 Jan 19;7:10326. doi: 10.1038/ncomms10326. Nat Commun. 2016. PMID: 26783717 Free PMC article. - Dutch population structure across space, time and GWAS design.
Byrne RP, van Rheenen W; Project MinE ALS GWAS Consortium; van den Berg LH, Veldink JH, McLaughlin RL. Byrne RP, et al. Nat Commun. 2020 Sep 11;11(1):4556. doi: 10.1038/s41467-020-18418-4. Nat Commun. 2020. PMID: 32917883 Free PMC article. - SNP and haplotype variation in the human genome.
Salisbury BA, Pungliya M, Choi JY, Jiang R, Sun XJ, Stephens JC. Salisbury BA, et al. Mutat Res. 2003 May 15;526(1-2):53-61. doi: 10.1016/s0027-5107(03)00014-9. Mutat Res. 2003. PMID: 12714183 Review.
Cited by
- Human genetic structure in Northwest France provides new insights into West European historical demography.
Alves I, Giemza J, Blum MGB, Bernhardsson C, Chatel S, Karakachoff M, Saint Pierre A, Herzig AF, Olaso R, Monteil M, Gallien V, Cabot E, Svensson E, Bacq D, Baron E, Berthelier C, Besse C, Blanché H, Bocher O, Boland A, Bonnaud S, Charpentier E, Dandine-Roulland C, Férec C, Fruchet C, Lecointe S, Le Floch E, Ludwig TE, Marenne G, Meyer V, Quellery E, Racimo F, Rouault K, Sandron F, Schott JJ, Velo-Suarez L, Violleau J, Willerslev E, Coativy Y, Jézéquel M, Le Bris D, Nicolas C, Pailler Y, Goldberg M, Zins M, Le Marec H, Jakobsson M, Darlu P, Génin E, Deleuze JF, Redon R, Dina C. Alves I, et al. Nat Commun. 2024 Aug 7;15(1):6710. doi: 10.1038/s41467-024-51087-1. Nat Commun. 2024. PMID: 39112481 Free PMC article. - Recent Historical Migrations Have Shaped the Gene Pool of Arabs and Berbers in North Africa.
Arauna LR, Mendoza-Revilla J, Mas-Sandoval A, Izaabel H, Bekada A, Benhamamouch S, Fadhlaoui-Zid K, Zalloua P, Hellenthal G, Comas D. Arauna LR, et al. Mol Biol Evol. 2017 Feb 1;34(2):318-329. doi: 10.1093/molbev/msw218. Mol Biol Evol. 2017. PMID: 27744413 Free PMC article. - Millennium-old pathogenic Mendelian mutation discovery for multiple osteochondromas from a Gaelic Medieval graveyard.
Jackson I, Mattiangeli V, Cassidy LM, Murphy E, Bradley DG. Jackson I, et al. Eur J Hum Genet. 2023 Feb;31(2):248-251. doi: 10.1038/s41431-022-01219-2. Epub 2022 Nov 28. Eur J Hum Genet. 2023. PMID: 36443465 Free PMC article. - Regional surnames and genetic structure in Great Britain.
Kandt J, Cheshire JA, Longley PA. Kandt J, et al. Trans Inst Br Geogr. 2016 Oct;41(4):554-569. doi: 10.1111/tran.12131. Epub 2016 Jul 7. Trans Inst Br Geogr. 2016. PMID: 27708455 Free PMC article. - Evidence of Polygenic Adaptation to High Altitude from Tibetan and Sherpa Genomes.
Gnecchi-Ruscone GA, Abondio P, De Fanti S, Sarno S, Sherpa MG, Sherpa PT, Marinelli G, Natali L, Di Marcello M, Peluzzi D, Luiselli D, Pettener D, Sazzini M. Gnecchi-Ruscone GA, et al. Genome Biol Evol. 2018 Nov 1;10(11):2919-2930. doi: 10.1093/gbe/evy233. Genome Biol Evol. 2018. PMID: 30335146 Free PMC article.
References
- Cardon LR, Bell JI. Association study designs for complex diseases. Nat. Rev. Genet. 2001;2:91–99. - PubMed
- Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nat. Genet. 2004;36:512–517. - PubMed
- Cavalli-Sforza LL, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton University Press; 1994.
- Quintana-Murci L, et al. Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat. Genet. 1999;23:437–441. - PubMed
ADDITIONAL REFERENCES FOR METHODS
- Bodmer JG, Eriksson AW, Forsius H, Nevanlinna HR, Workman PL, Norio RK. Popul. Struct. Genet. Disord. Academic Press; 1980. pp. 211–238.
Publication types
MeSH terms
Grants and funding
- 075491/Z/04/Z/WT_/Wellcome Trust/United Kingdom
- 098387/WT_/Wellcome Trust/United Kingdom
- 095552/Z/11/Z/WT_/Wellcome Trust/United Kingdom
- 072974/Z/03/Z/WT_/Wellcome Trust/United Kingdom
- MC_UU_12013/1/MRC_/Medical Research Council/United Kingdom
- 104125/WT_/Wellcome Trust/United Kingdom
- 075491/WT_/Wellcome Trust/United Kingdom
- 075491/Z/04/B/WT_/Wellcome Trust/United Kingdom
- 090532/WT_/Wellcome Trust/United Kingdom
- 088262/WT_/Wellcome Trust/United Kingdom
- 090532/Z/09/Z/WT_/Wellcome Trust/United Kingdom
- 098387/Z/12/Z/WT_/Wellcome Trust/United Kingdom
- MR/M501608/1/MRC_/Medical Research Council/United Kingdom
- 085475/Z/08/Z/WT_/Wellcome Trust/United Kingdom
- 084818/Z/08/Z/WT_/Wellcome Trust/United Kingdom
- 095552/WT_/Wellcome Trust/United Kingdom
- 098386/Z/12/Z/WT_/Wellcome Trust/United Kingdom
- R01 NS049477/NS/NINDS NIH HHS/United States
- 072974/WT_/Wellcome Trust/United Kingdom
- 085475DONNELLY/WT_/Wellcome Trust/United Kingdom
- 088262/Z/09/Z/WT_/Wellcome Trust/United Kingdom
- 084818/WT_/Wellcome Trust/United Kingdom
- 075491/Z/04/A/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 098386/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources