Two distinct modes for propagation of histone PTMs across the cell cycle - PubMed (original) (raw)
Two distinct modes for propagation of histone PTMs across the cell cycle
Constance Alabert et al. Genes Dev. 2015.
Abstract
Epigenetic states defined by chromatin can be maintained through mitotic cell division. However, it remains unknown how histone-based information is transmitted. Here we combine nascent chromatin capture (NCC) and triple-SILAC (stable isotope labeling with amino acids in cell culture) labeling to track histone modifications and histone variants during DNA replication and across the cell cycle. We show that post-translational modifications (PTMs) are transmitted with parental histones to newly replicated DNA. Di- and trimethylation marks are diluted twofold upon DNA replication, as a consequence of new histone deposition. Importantly, within one cell cycle, all PTMs are restored. In general, new histones are modified to mirror the parental histones. However, H3K9 trimethylation (H3K9me3) and H3K27me3 are propagated by continuous modification of parental and new histones because the establishment of these marks extends over several cell generations. Together, our results reveal how histone marks propagate and demonstrate that chromatin states oscillate within the cell cycle.
Keywords: DNA replication; cell cycle; epigenetics; histone post-translational modifications; histone recycling; histone variants.
© 2015 Alabert et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
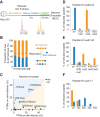
Figure 1.
Histones are recycled together with their modifications on newly replicated DNA. (A) Experimental design. Cells were synchronized by thymidine and released in heavy SILAC medium. Newly replicated DNA was labeled with b-dUTP and isolated on streptavidin beads by NCC (Supplemental Fig. S1A). (B) Proportion of new and old histones on nascent chromatin. The dotted line indicates theoretical values. (C) Overview of how histone PTMs in nascent chromatin distribute on new and old histones. The mean of nine independent biological experiments is shown (n = 9). Old histones are mainly methylated (me), while new ones are acetylated (ac). Only shared marks are labeled. For values for all identified PTMs, see Supplemental Figure S1C. (D_–_F) Modifications on new (blue) and old (orange) histone H3 and H4 in nascent chromatin shown as a percentage of heavy or light peptides, respectively. The H4 amino acid 4–17 (aa4–17) peptide contains K5, K8, K12, and K16. (Un) Unmodified; (ac) acetylated; (ac2) diacetylated; (ac3) triacetylated; (ac4) quadriacetylated; (me1) monomethylated; (me2) dimethylated; (me3) trimethylated. K27/K36me1 is resolved in Supplemental Figure S1, E and F.
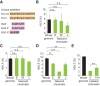
Figure 2.
Recycling of histone variants at replication forks. Experimental design as in Figure 1A except that nascent chromatin was collected in early (E), mid- (M), and late (L) S phase (Supplemental Fig. S3A). (A) Unique peptides used to differentiate canonical histones H3.1 and H3.2 from the H3.3 variant as well as histone H2A from H2A.X and H2A.Z variants. (B) Enrichment of H3.3 relative to total H3 (H3.3+H3.1/2). (C,D) Enrichment of H2A.X and H2A.Z relative to total H2A (H2A+H2A.Z+H2AX). (E) Enrichment of H2A.Z in labeled chromatin at 0 h (nascent; mid-S phase) and 10 h later (see Supplemental Fig. S4A for experimental design). Error bars indicate SD. 3 < n < 9. Unpaired _t_-test: (***) P < 0.001; (**) P < 0.05; (n.s.) nonsignificant.
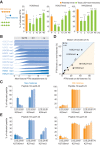
Figure 3.
Propagation of histone PTMs across the cell cycle. Histone PTM propagation was determined by NCC coupled with double- and triple-SILAC labeling in a time-course analysis (Supplemental Figs. S4A, S5A). (A) Kinetics of PTM restoration. The total PTM level on replicated DNA (green) was compared with the PTM level of the old histones alone (P; orange), which, in nascent chromatin, represents the parental chromatin state that should to be reached by new and old histones together at the next round of replication, where they (new+old) constitute the parental histones of the daughter cells. After 24 h, the total PTM level of new+old histones (24 h; green) mirrors the parental histones of the previous round of replication (P; orange), showing that the chromatin state is propagated. Error bars indicate SD. n ≥ 3. Unpaired _t_-test identified differences from the parental state. A combination of double- and triple-SILAC time-course data is shown. (B) Kinetics of PTM establishment on new histones. (C) Dynamics of H4K20 methylation on new and old histones. Error bars indicate SD. n ≥ 3. (D) Overview of how histone PTMs in mature chromatin (24 h) distribute on new and old histones (n = 3). Old and new histones are identical except for the indicated marks. For values for all identified PTMs throughout the time course, C and E, and Supplemental Figure S6, A and B. (E) Dynamics of H3K27 methylation on new and old histones. Error bars indicate SD. n ≥ 3.
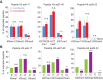
Figure 4.
Cell cycle control of histone PTM levels. (A) After b-dUTP labeling, cells were treated with (+) or without (−) nocodazole for an additional 10, 16, or 18.5 h (Supplemental Fig. S7A). The mean of three time points is shown, with error bars indicating SD. n ≥ 3. Unpaired _t_-test: (****) P < 0.0001; (*) P < 0.05. (B) Analysis of histone PTM levels in exponentially growing (green) and contact-inhibited (purple) primary TIG-3 fibroblasts. Mean is shown, with error bars indicating SD. n = 3. Unpaired _t_-test: (****) P < 0.0001; (**) P < 0.01. For an outline of the experimental design, see Supplemental Figure S7C.
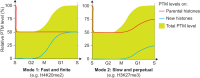
Figure 5.
Illustration of the two basic principles for histone PTM propagation. The contribution of new and parental histones (lines) to the total PTM level (green) is shown. (Mode 1) New histones acquire PTMs to become identical to the parental histones within one cell cycle (H3K9me1, H3K27/36me1, H3K36me2, H3K9me2, H3K27me2, H3K79me1, H3K79me2, H4K20me1, and H4K20me2); (Mode 2) propagation relies on progressive modification of both new and parental histones (H3K9me3 and H3K27me3).
Similar articles
- Accurate Recycling of Parental Histones Reproduces the Histone Modification Landscape during DNA Replication.
Reverón-Gómez N, González-Aguilera C, Stewart-Morgan KR, Petryk N, Flury V, Graziano S, Johansen JV, Jakobsen JS, Alabert C, Groth A. Reverón-Gómez N, et al. Mol Cell. 2018 Oct 18;72(2):239-249.e5. doi: 10.1016/j.molcel.2018.08.010. Epub 2018 Aug 23. Mol Cell. 2018. PMID: 30146316 Free PMC article. - Investigating Mitotic Inheritance of Histone Posttranslational Modifications by Triple pSILAC Coupled to Nascent Chromatin Capture.
Nakamura K, Groth A, Alabert C. Nakamura K, et al. Methods Mol Biol. 2022;2529:407-417. doi: 10.1007/978-1-0716-2481-4_17. Methods Mol Biol. 2022. PMID: 35733024 - SILAC-based proteomic analysis to dissect the "histone modification signature" of human breast cancer cells.
Cuomo A, Moretti S, Minucci S, Bonaldi T. Cuomo A, et al. Amino Acids. 2011 Jul;41(2):387-99. doi: 10.1007/s00726-010-0668-2. Epub 2010 Jul 9. Amino Acids. 2011. PMID: 20617350 - Restoring chromatin after replication: how new and old histone marks come together.
Jasencakova Z, Groth A. Jasencakova Z, et al. Semin Cell Dev Biol. 2010 Apr;21(2):231-7. doi: 10.1016/j.semcdb.2009.09.018. Epub 2009 Oct 6. Semin Cell Dev Biol. 2010. PMID: 19815085 Review. - Interpreting the language of histone and DNA modifications.
Rothbart SB, Strahl BD. Rothbart SB, et al. Biochim Biophys Acta. 2014 Aug;1839(8):627-43. doi: 10.1016/j.bbagrm.2014.03.001. Epub 2014 Mar 12. Biochim Biophys Acta. 2014. PMID: 24631868 Free PMC article. Review.
Cited by
- The Structural Determinants behind the Epigenetic Role of Histone Variants.
Cheema MS, Ausió J. Cheema MS, et al. Genes (Basel). 2015 Jul 23;6(3):685-713. doi: 10.3390/genes6030685. Genes (Basel). 2015. PMID: 26213973 Free PMC article. Review. - Interactions With Histone H3 & Tools to Study Them.
Scott WA, Campos EI. Scott WA, et al. Front Cell Dev Biol. 2020 Jul 31;8:701. doi: 10.3389/fcell.2020.00701. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32850821 Free PMC article. Review. - Dynamics of Nucleosome Positioning Maturation following Genomic Replication.
Vasseur P, Tonazzini S, Ziane R, Camasses A, Rando OJ, Radman-Livaja M. Vasseur P, et al. Cell Rep. 2016 Sep 6;16(10):2651-2665. doi: 10.1016/j.celrep.2016.07.083. Epub 2016 Aug 25. Cell Rep. 2016. PMID: 27568571 Free PMC article. - Epigenetic Regulation by Polycomb Complexes from Drosophila to Human and Its Relation to Communicable Disease Pathogenesis.
Scholl A, De S. Scholl A, et al. Int J Mol Sci. 2022 Oct 14;23(20):12285. doi: 10.3390/ijms232012285. Int J Mol Sci. 2022. PMID: 36293135 Free PMC article. Review. - Re-establishment of nucleosome occupancy during double-strand break repair in budding yeast.
Tsabar M, Hicks WM, Tsaponina O, Haber JE. Tsabar M, et al. DNA Repair (Amst). 2016 Nov;47:21-29. doi: 10.1016/j.dnarep.2016.09.005. Epub 2016 Sep 28. DNA Repair (Amst). 2016. PMID: 27720308 Free PMC article.
References
- Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, et al.1999. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J 18: 1923–1938. - PMC - PubMed
- Alabert C, Groth A. 2012. Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol 13: 153–167. - PubMed
- Alabert C, Bukowski-Wills JC, Lee SB, Kustatscher G, Nakamura K, de Lima Alves F, Menard P, Mejlvang J, Rappsilber J, Groth A. 2014. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat Cell Biol 16: 281–293. - PMC - PubMed
- Annunziato AT. 2005. Split decision: what happens to nucleosomes during DNA replication? J Biol Chem 280: 12065–12068. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous