Aerobic glycolysis tunes YAP/TAZ transcriptional activity - PubMed (original) (raw)
. 2015 May 12;34(10):1349-70.
doi: 10.15252/embj.201490379. Epub 2015 Mar 21.
Giulia Santinon 1, Arianna Pocaterra 1, Mariaceleste Aragona 1, Silvia Bresolin 2, Mattia Forcato 3, Daniela Grifoni 4, Annalisa Pession 4, Francesca Zanconato 1, Giulia Guzzo 5, Silvio Bicciato 3, Sirio Dupont 6
Affiliations
- PMID: 25796446
- PMCID: PMC4491996
- DOI: 10.15252/embj.201490379
Aerobic glycolysis tunes YAP/TAZ transcriptional activity
Elena Enzo et al. EMBO J. 2015.
Abstract
Increased glucose metabolism and reprogramming toward aerobic glycolysis are a hallmark of cancer cells, meeting their metabolic needs for sustained cell proliferation. Metabolic reprogramming is usually considered as a downstream consequence of tumor development and oncogene activation; growing evidence indicates, however, that metabolism on its turn can support oncogenic signaling to foster tumor malignancy. Here, we explored how glucose metabolism regulates gene transcription and found an unexpected link with YAP/TAZ, key transcription factors regulating organ growth, tumor cell proliferation and aggressiveness. When cells actively incorporate glucose and route it through glycolysis, YAP/TAZ are fully active; when glucose metabolism is blocked, or glycolysis is reduced, YAP/TAZ transcriptional activity is decreased. Accordingly, glycolysis is required to sustain YAP/TAZ pro-tumorigenic functions, and YAP/TAZ are required for the full deployment of glucose growth-promoting activity. Mechanistically we found that phosphofructokinase (PFK1), the enzyme regulating the first committed step of glycolysis, binds the YAP/TAZ transcriptional cofactors TEADs and promotes their functional and biochemical cooperation with YAP/TAZ. Strikingly, this regulation is conserved in Drosophila, where phosphofructokinase is required for tissue overgrowth promoted by Yki, the fly homologue of YAP. Moreover, gene expression regulated by glucose metabolism in breast cancer cells is strongly associated in a large dataset of primary human mammary tumors with YAP/TAZ activation and with the progression toward more advanced and malignant stages. These findings suggest that aerobic glycolysis endows cancer cells with particular metabolic properties and at the same time sustains transcription factors with potent pro-tumorigenic activities such as YAP/TAZ.
Keywords: Hippo pathway; TEAD; YAP/TAZ; aerobic glycolysis; glucose metabolism.
© 2015 The Authors.
Figures
Figure 1
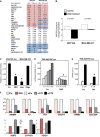
Glucose metabolism regulates YAP/TAZ transcriptional activity
- Over-representation analysis was performed with gene signatures highlighting activation of specific pathways using gene set enrichment analysis (GSEA) on microarray data obtained from MCF10A or MDA-MB-231 mammary cells untreated or treated with 2-deoxy-glucose (2DG, 50 mM) to inhibit glucose metabolism. The normalized enrichment score (NES) is the primary statistic for examining GSEA results; a positive NES (highlighted in red) indicates signatures expressed more in control cells than upon 2DG treatment (i.e. signatures activated when glucose metabolism is active); a negative NES (highlighted in blue) indicates signatures expressed more upon 2DG treatment. The false discovery rate (FDR) is the estimated probability that a gene set with a given NES represents a false positive; we considered signatures to be significantly enriched at FDR < 0.05. Gene expression data have been obtained from n = 4 biological replicates for each condition. See Supplementary Table S1 for a GSEA analysis including also Biocarta gene sets.
- 2DG treatment downregulates the overall levels of the ‘YAP/TAZ’ gene signature used in (A) as calculated from microarray data of cells untreated (white bars) or treated with 2DG (black bars). See Materials and Methods for details on the statistical methods to quantify average signature expression. Data are shown as mean ± standard error of the mean (SEM). Of note, in this analysis, the basal levels of YAP/TAZ target genes were higher in the cell line displaying higher glycolysis/respiration ratio, that is, in MDA-MB-231 cells (Supplementary Fig S1B).
- Luciferase assay in MDA-MB-231 breast cancer cells transfected with the synthetic YAP/TAZ reporter 8XGTIIC-lux. Starting on the day after DNA transfection, cells were treated for 24 h with the indicated small-molecule inhibitors to block glucose metabolism (50 mM 2DG; 1 mM lonidamine, Loni) or with an inhibitor of the mitochondrial respiratory chain (1 μM oligomycin, Oligo). Activity of the reporter is normalized to cotransfected CMV-lacZ and expressed relative to the cells treated with vehicle only (Co.). See Supplementary Fig S1E–K for controls on the specificity of 2DG treatment and similar results obtained in Hs578T and HepG2 cells. Representative results of a single experiment with n = 2 biological replicates; four independent experiments were consistent.
- Luciferase assay in MDA-MB-231 cells bearing a stably integrated TRE-8XGTIIC-lux reporter, whose transcription can be released following doxycycline treatment to visualize early YAP/TAZ responses (see Supplementary Fig S1N for controls). Control cells (Co.) were left unstimulated (0) or supplemented with doxycycline (4, 6, 8 and 10 h of treatment) to release YAP/TAZ-dependent transcription. 2DG (100 mM) was added together with doxycycline to acutely block glucose metabolism. See Supplementary Fig S1P–R for similar results obtained in MCF10A-MII cells. Representative results of a single experiment with n = 2 biological replicates; three independent experiments were consistent.
- Luciferase assay was carried out as in (D), by removing glucose from the culture medium at the moment of doxycycline supplementation (−Glu). Cells were harvested 24 h after treatment. See Supplementary Fig S1O and R for similar results obtained in HepG2 and MCF10A-MII cells. Representative results of a single experiment with n = 2 biological replicates; three independent experiments were consistent.
- YAP/TAZ are required for transcription of 2DG-regulated genes. qPCR for endogenous target genes in MDA-MB-231 cells treated with water (Co.) or with 2DG or transfected with the indicated siRNAs: control (siCo.), YAP/TAZ mix #1 (siYT1), YAP/TAZ mix #2 (siYT2). Expression levels were calculated relative to GAPDH and are given relative to Co. cells (arbitrarily set to 1). Genes were selected among the probes commonly regulated in microarray profiling (see Supplementary Table S3). Note how both 2DG-induced and 2DG-inhibited genes were coherently regulated by YAP/TAZ knockdown. See Supplementary Fig S1S for other targets and controls, and Supplementary Fig S1T for similar results in Hs578T cells. n = 4 biological replicates from two independent experiments. All differences had _P_-value < 0.01.
Data information: Unless indicated otherwise, error bars represent mean ± SD. *_P_-value < 0.01 relative to control.
Figure 2
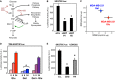
Glycolysis sustains YAP/TAZ activity
- A simplified scheme indicating the main metabolic routes followed by glucose, the key intermediates and enzymes involved, and the inhibitors used in this study. Only the pathways and enzymes discussed in the text are shown here for simplicity. G6P: glucose-6-phosphate; F6P: fructose-6-phosphate; F1,6P: fructose-1,6-bisphosphate; F2,6P: fructose-2,6-bisphosphate; GlcNAc: N-acetyl glucosamine; HK: hexokinase; GPI: phosphoglucoisomerase; PFK1: 6-phosphofructo-1-kinase; PFKFB3: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3. Lonidamine (Loni.) inhibits HK (Tennant et al, 2010); 2DG inhibits both HK and GPI (Wick et al, ; Tennant et al, 2010); DON and AZS inhibit the enzyme mediating the first step of the hexosamine pathway (Wellen et al, ; Ostrowski & van Aalten, ; Onodera et al, 2014). The green arrow indicates the agonistic effect of F2,6P on PFK1. Dashed arrows indicate downstream intermediates or metabolic pathways.
- Phosphoglucoisomerase (GPI) is required for YAP/TAZ activity. Luciferase assay in MDA-MB-231 cells transfected with the indicated siRNAs. See Supplementary Fig S2A for validation of siRNA efficiency. Representative results of a single experiment with n = 2 biological replicates; three independent experiments were consistent.
- The plot indicates basal oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) of TRE-8XGTIIC-lux MDA-MB-231 cells grown in glucose (red, Glu) or 10 mM galactose (blue, Gal). As expected, galactose induces a metabolic shift from aerobic glycolysis to oxidative phosphorylation compared to glucose. See Supplementary Fig S2D–F for detailed OCR and ECAR traces. Representative results of a single experiment with n = 5 biological replicates; two independent experiments were consistent.
- Comparison of YAP/TAZ activity in MDA-MB-231 cells bearing a stably integrated TRE-8XGTIIC-lux reporter and grown in glucose (red, Glu), in galactose to induce a shift toward oxidative respiration (blue, Gal), or shifted back to glucose during doxycycline treatment (blue bars with red stripes, Gal + Glu). Cells were treated with doxycycline to release YAP/TAZ-dependent luciferase transcription for 8 or 24 h. Galactose-fed cells display reduced glycolysis and downregulate YAP/TAZ activity. Glucose rapidly reactivates glycolysis (Supplementary Fig S2F) and YAP/TAZ activity (Gal + Glu). Representative results of a single experiment with n = 2 biological replicates; three independent experiments were consistent.
- Luciferase assay in UOK262 kidney cancer cells, bearing mutation of the fumarate hydratase (FH) enzyme of the tricarboxylic acid cycle (TCA). FH-reconstituted cells (gray bars) display a reduction of aerobic glycolysis and increased respiration (Yang et al, 2013). 2DG treatment (12 mM) of parental cells serves as a positive control for inhibition of the glycolysis–YAP/TAZ axis in parental cells. Representative results of a single experiment with n = 2 biological replicates; two independent experiments were consistent.
Data information: Throughout the figure, error bars represent mean ± SD. *_P_-value < 0.01.
Figure 3
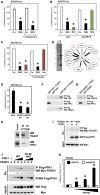
Phosphofructokinase regulates YAP/TAZ transcriptional activity and interacts with TEADs
- Luciferase assay in MDA-MB-231 cells treated for 24 h with 2DG (black bars) and/or with compound-C (30 μM), an established inhibitor of AMPK. The same dose of compound-C is sufficient to prevent AMPK activation by 2DG (see Supplementary Fig S3C), but not YAP/TAZ inhibition. Representative results of a single experiment with n = 2 biological replicates; three independent experiments were consistent. See Supplementary Fig S3D and E for similar results with AMPKa1/2 silencing.
- Luciferase assay in cells transfected with control (siCo.) or with an established LATS1/2 siRNA mix (siLATS1/2) (Aragona et al, 2013) and then either treated with 2DG (black bars) or transfected with NF2 expression plasmid to specifically activate the Hippo pathway (green bars). Depletion of LATS1/2 blocked the inhibitory effect of overexpressed NF2, but not of 2DG. Similar results were obtained with an independent mix of LATS1/2 siRNA (data not shown). Representative results of a single experiment with n = 2 biological replicates; two independent experiments were consistent.
- Inhibition of glycolysis could potentially deplete cells of acetyl-CoA, the main precursor for mevalonate, and mevalonate is required for YAP/TAZ activity by regulating RHO GTPases (Sorrentino et al, ; Wang et al, 2014). Cells were transfected with the 8XGTIIC-lux YAP/TAZ reporter and treated with 2DG (black bars) or with cerivastatin (3 μM, red bars), an inhibitor of the mevalonate pathway at the level of HMG-CoA reductase. Adding back mevalonate in the culture medium (+ mevalonate, 1 mM) rescues YAP/TAZ inhibition from cerivastatin, but not from 2DG. Representative results of a single experiment with n = 2 biological replicates; two independent experiments were consistent.
- Proteomic analysis of YAP-binding partners reveals interaction with phosphofructokinase (PFK1). Flag-tagged YAP-5SA stably expressed in MCF10A and MDA-MB-231 cells was immunoprecipitated, and associated proteins identified using mass spectrometry. Left panel: silver staining of the purified proteins in representative control (Co.) or YAP immunopurifications. Molecular weight markers are indicated. The asterisk indicates the band corresponding to YAP. Right scheme: The thickness of the lines connecting YAP to its partners is proportional to the number of peptides isolated for each partner. Black proteins (known YAP partners) and PFK1 (in red) were isolated in both cell lines; gray proteins are known regulators of YAP that were only purified from MCF10A cells. See Supplementary Table S4 for a complete list of the identified peptides.
- Luciferase assay in MDA-MB-231 cells transfected with control (siCo.) or two independent PFK1 siRNAs (siPFK1 #1, #2). Representative results of a single experiment with n = 2 biological replicates; four independent experiments were consistent. See Supplementary Fig S3H for validation of PFK1 siRNAs and Supplementary Fig S3I for similar results on CTGF-lux.
- In vitro pull-down assay with purified FLAG-PFK1 and recombinant GST-YAP. GST-YAP was incubated with (first lane) or without (second lane) FLAG-PFK1; as positive control, GST-YAP was incubated with purified FLAG-TEAD1 (right-most lane). Proteins were then subjected to anti-FLAG immunoprecipitation, and purified complexes were probed for coprecipitation of GST-YAP (anti-YAP immunoblot).
- In vitro pull-down assay with purified FLAG-PFK1 and recombinant GST-TEAD4. GST-TEAD4 was incubated with (first lane) or without (second lane) FLAG-PFK1. Proteins were then subjected to anti-FLAG immunoprecipitation, and purified complexes were probed for coprecipitation of GST-TEAD4 (anti-TEAD4 immunoblot).
- MDA-MB-231 cell lysates were immunoprecipitated with anti-TEAD1 antibody, and the precipitating proteins were probed for TEAD1 or PFK1. Immunoprecipitation with an unrelated IgG serves as negative control. Of note, this interaction is in line with the requirement of TEAD1 and TEAD4 for YAP/TAZ activity in our cellular systems (Supplementary Fig S3L and M).
- Lysates from HEK293 cells transfected with the indicated proteins were subjected to anti-FLAG-PFK1 immunoprecipitation, and purified complexes were probed for coprecipitation of MYC-TEAD4. Mutation of a key amino acid required for interaction between TEAD4 and YAP/TAZ (Y429H) did not interfere with PFK1 interaction.
- Mutation of the fructose-2,6-P allosteric site of PFK1 negatively regulates its interaction with TEAD4. HEK293 cells were transfected with MYC-TEAD4 and increasing doses of wild-type (WT) or mutated (F2,6P-mut) FLAG-PFK1 plasmids; cell extracts were immunoprecipitated with anti-FLAG, and the coprecipitating MYC-TEAD4 protein was detected by Western blotting. Immunoprecipitation in the absence of FLAG-PFK1 (lane 1) serves as a negative control. Quantifications of the TEAD4/PFK1 ratio are provided, relative to lane 2.
- Luciferase assay in HEK293 cells transfected with 8XGTIIC-lux reporter (black bars) or with the reporter deleted of the TEAD-binding sites (delta8XGT), and with increasing doses of PFKFB3 expression plasmid. PFKFB3 converts fructose-6-P into fructose-2,6-P, a potent allosteric activator of PFK1 (Sola-Penna et al, 2010). Representative results of a single experiment with n = 2 biological replicates; three independent experiments were consistent. See Supplementary Fig S3P for controls of the delta8XGT reporter.
Data information: Throughout the figure, error bars represent mean ± SD. *_P_-value < 0.01. Source data are available online for this figure.
Figure 4
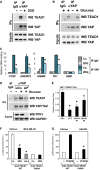
Glucose metabolism regulates the interaction between YAP/TAZ and TEADs
- Extracts of MDA-MB-231 cells treated for 24 h with vehicle (−) or with 2DG (+) were subjected to anti-YAP immunoprecipitation; coprecipitating proteins were then analyzed by Western blotting to detect TEAD1 interaction. Immunoprecipitation with an unrelated IgG serves as a negative control. Similar results were obtained in other cell lines, or in MDA-MB-231 cells by using a different anti-YAP/TAZ antibody (Supplementary Fig S4B–E).
- Coimmunoprecipitations from extracts of MCF10A cells released from contact inhibition by seeding them at low confluence for 36 h with glucose (+, lane 2), without glucose (−, lane 3), or cultured without glucose and then refed of glucose (+, lane 4). Immunoprecipitation with an unrelated IgG (lane 1) serves as a negative control. Similar results were obtained in HepG2 (Supplementary Fig S4F).
- Chromatin immunoprecipitation of MCF10A cells untreated (Co.) or treated with 100 mM 2DG for 24 h. Fragmented chromatin from each experimental condition was immunoprecipitated with control IgG or anti-YAP antibodies and subjected to qPCR to detect the TEAD-binding regions present in the CTGF, ANKRD1, HMMR and TK1 promoters. Amplification of Hemoglobin beta (HBB) serves as a negative control. See Supplementary Fig S4G for similar results in MDA-MB-231 cells. CTGF, ANKRD1 and RHAMM are known targets of YAP/TAZ; TK1 was included in the analysis because it is jointly regulated by glucose and YAP/TAZ (see Fig1F), and its proximal promoter contains two TEAD-binding sites that respond to YAP/TAZ activity (see Supplementary Fig S4H) and to glycolysis coherently (Supplementary Fig S4I). Values in control samples with control IgG were arbitrarily set to 1, and the other values are relative to this (see Materials and Methods). Data are shown as the mean ± SD of two independent experiments.
- Immunoprecipitation of YAP and TEAD1 is reinforced upon glucose supplementation (+), and this requires endogenous PFK1 levels (compare siCo. with siP siRNA transfected extracts).
- In situ interaction of YAP and TEAD1 is regulated by PFK1 by proximity ligation assay (PLA). Depletion of YAP/TAZ (siYT) or PFK1 with two independent siRNAs (siPFK1 #1, #2) reduced the number of nuclear YAP/TEAD1 dots relative to cells transfected with control siRNA (siCo.), suggesting PFK1 is required to stabilize YAP/TEAD1 interaction. See Supplementary Fig S4J for representative pictures of the PLA stainings.
- Luciferase assay in MDA-MB-231 cells transfected with UAS-lux reporter and expression plasmids encoding for in-frame fusions of the GAL4 DNA-binding domain with TEAD1, wild-type (WT) or unable to interact with YAP (Y406A mutant). WT TEAD1 can recruit YAP/TAZ and efficiently promote transcription, while Y406A TEAD1 can only sustain basal transcription (Li et al, 2010). Treatment with 2DG (black bars) inhibited transcription induced by WT TEAD1 but not of the Y406A mutant, in keeping with the observation that 2DG regulates YAP/TEAD1 interaction. Latrunculin A treatment (Lat.A) serves as a positive control for inhibition of YAP/TAZ. Representative results of a single experiment with n = 2 biological replicates; two independent experiments were consistent.
- Luciferase assay in HEK293 cells transfected as in (F). Cotransfection of PFKFB3, an activator of PFK1 activity, fosters the activity of TEAD1 only when it is able to interact with YAP. Representative results of a single experiment with n = 2 biological replicates; two independent experiments were consistent.
Data information: Throughout the figure, error bars represent mean ± SD. *_P_-value < 0.01. Source data are available online for this figure.
Figure 5
Interplay of glycolysis, PFK1 and YAP/TAZ in cancer cell growth
- A Mammosphere assay with MCF10A-MII cells. Retroviral expression of an activated form of TAZ (S89A mutant) increases the efficiency of primary mammosphere formation compared to parental cells (empty-vector transduced cells). Treatment of TAZ-expressing cells with 2DG (15 mM), or depletion of PFK1 (siP) or GPI (siG), impairs the mammosphere-promoting ability of TAZ. See Supplementary Fig S5A–C for secondary mammospheres and representative pictures. Representative results of a single experiment with n = 4 biological replicates; two independent experiments were consistent.
- B Depletion of PFK1 (siP) impairs the colony-forming ability of MDA-MB-231 cells in soft agar, recapitulating the requirement for YAP/TAZ (siYT). Representative results of a single experiment with n = 2 biological replicates; two independent experiments were consistent.
- C Expression of an activated form of YAP (5SA) strongly promotes the growth of MDA-MB-231 colonies in soft agar, and this is inhibited by 2DG treatment (3 mM). Each box signifies the upper and lower quartiles of data (colony size), while the whiskers extend to the minimum and maximum data points. On the right: representative pictures of colonies growing from 5SA-YAP-expressing cells, treated with vehicle or with 2DG. Representative results of a single experiment with n = 2 biological replicates; four independent experiments were consistent.
- D MCF10A cells were seeded at high density for 48 h, leading to YAP/TAZ inhibition and growth arrest (High); scratching the monolayer locally enables cell spreading and activates YAP/TAZ, thus inducing cell proliferation (Wound) (Zhao et al, ; Aragona et al, 2013). Overnight treatment of cells with 2DG (15 mM) inhibited such YAP/TAZ-induced proliferation. The graph reports the quantification of proliferating cells in the indicated areas, without or with 2DG treatment. To count cells abutting the wound, we arbitrarily set a 100-μm distance from the wound. See Supplementary Fig S5D for representative pictures of a wounded area. Representative results of a single experiment with n = 2 biological replicates (> 700 cells/replicate); three independent experiments were consistent.
- E Clonal expansion induced by overexpression of the YAP homologue Yki in the Drosophila wing imaginal disk is restricted by phosphofructokinase (Pfk) RNAi. Panels on the left show pictures of wing imaginal disks bearing clones of cells (marked by GFP) with mutation of the lethal giant larvae tumor suppressor gene (_lgl_−) and overexpression of Yki (yki over), induced by the MARCM technique (Lee & Luo, 2001). The dotted line indicates the outline of the disks. In this genetic setup, the survival of clones within the wing pouch, that is, in the distal region of the wing disk, is strictly dependent on Yki activation (Grzeschik et al, ; Menendez et al, ; Khan et al, 2013). Upon downregulation of phosphofructokinase, the growth of _lgl_−; yki over clones was inhibited, as shown by quantification of clone area (****P < 0.0001, unpaired _t_-test). n = 34 disks for each genotype. Scale bars 80 μm.
- F Pfk silencing downregulates the Yki target gene DIAP1 in _lgl_−; yki over clones. Panels show whole-mount immunostainings for DIAP1 protein levels, a hallmark of Yki transcriptional activity (Huang et al, 2005), on wing imaginal disks of the indicated phenotypes, as in (E). GFP identifies mutant cells, growing within an otherwise wild-type tissue. DIAP1 is autonomously upregulated in _lgl_−; yki over clones, while it appears downregulated upon Pfk RNAi. This is consistent with a role for Pfk in regulating Yki transcriptional activity. See Supplementary Fig S5E for lower magnifications of the same wing disks. See Supplementary Fig S5F and G for similar results obtained with dMyc, another target of Yki (Neto-Silva et al, ; Ziosi et al, 2010). Scale bars, 20 μm.
- G MDA-MB-231 cells were growth-inhibited by glucose withdrawal for 48 h (−Glu), and then proliferation was induced by supplementing glucose in the medium for 24 h (+Glu). Culture medium was without glutamine to specifically measure glucose-dependent growth. Quantification of proliferation, as measured by BrdU incorporation, indicates that cells depleted of YAP/TAZ (siYT #1) are unable to efficiently restart proliferation in response to glucose compared to cells transfected with control siRNA (siCo.). Similar results were obtained with an independent YAP/TAZ siRNA mix (not shown). Representative results of a single experiment with n = 2 biological replicates (> 1,000 cells/replicate); three independent experiments were consistent.
- H, I Clonogenic assay with UOK262 cells. Parental cells (black bars) are highly glycolytic, while their FH-reconstituted counterpart (gray bars) has reduced glycolysis as they can efficiently perform mitochondrial respiration (Yang et al, 2013). Cells were seeded at clonogenic density and grown in the presence of titrated doses of 2DG (0.25, 0.5, 1 mM) to inhibit glucose metabolism (H) or in the presence of VP (0.3, 1, 3 μM) to inhibit the cooperation between YAP/TAZ and TEADs (Liu-Chittenden et al, 2012) (I). Graphs show the quantification of colonies after 10 days, relative to untreated cells. UOK262 cells are more sensitive than UOK262-FH to 2DG; UOK262 cells are also more sensitive to small-molecule inhibition of YAP/TAZ, in keeping with higher YAP/TAZ activity (shown above). Representative results of a single experiment with n = 3 biological replicates; two independent experiments were consistent.
Data information: Throughout the figure, error bars represent mean ± SD. *_P_-value < 0.01.
Figure 6
YAP/TAZ activity is enhanced in primary human breast cancers displaying high levels of a gene signature associated to glycolysis
- A Scatter plot (gray dots) and linear regression (red line, slope 0.532) of standardized expression values indicate a positive correlation between a gene signature experimentally associated with active glucose metabolism (glucose signature) and a gene signature denoting YAP/TAZ activity, in a metadataset collecting n = 3,661 primary human breast cancers (see Materials and Methods). The glucose signature is composed of the genes downregulated upon 2DG treatment, that is, requiring active glucose metabolism for their transcription, both in MCF10A and in MDA-MB-231 cells (see Supplementary Table S5). Pearson ρ quantifies the linear dependence between the levels of the two signatures. The coefficient of determination is _r_2 = 0.731, _P_-value < 0.0001.
- B Primary human breast cancers of the metadataset were stratified according to high or low glucose signature score, and then, the levels of the YAP/TAZ signature score were determined in the two groups (see Materials and Methods for details on the statistical methods to quantify scores). YAP/TAZ activity is significantly higher in tumors with high levels of the glucose signature, as visualized by box-plot. The bottom and top of the box are the first and third quartiles, and the band inside the box is the median; whiskers represent 1st and 99th percentiles; values lower and greater are shown as circles (P < 0.0001, n = 3,661).
- C, D Primary human breast cancers of the metadataset were classified according to high or low glucose signature score, and then, the levels of the Staminal or stem tumorigenic signature scores, associated to normal and cancer mammary stem cells, were determined in the two groups. Gene expression associated to mammary stem cells is significantly higher in tumors with high levels of the glucose signature, as visualized by box-plot (P < 0.0001, n = 3,661).
- E Genes regulated by glucose metabolism (glucose signature) are elevated in G3 as compared to G1 grade mammary tumors of the metadataset (P < 0.0001; G1 versus G3 unpaired _t_-test). A similar behavior is observed by using the YAP/TAZ signature (Cordenonsi et al, 2011). See Materials and Methods for details on the statistical methods to quantify average signature expression. Data are shown as mean ± standard error of the mean (SEM).
- F Kaplan–Meier analysis representing the probability of metastasis-free survival in breast cancer patients from the metadataset stratified according to high or low glucose signature score. The log-rank test _P_-value reflects the significance of the association between high levels of the glucose signature score and shorter survival. A similar behavior is observed by using the YAP/TAZ signature (Cordenonsi et al, 2011).
- G Genes regulated by glucose metabolism but not by YAP/TAZ (glucose NOT YT signature, see Supplementary Table S5) are not expressed at higher levels in G3 grade mammary tumors of the metadataset. See Materials and Methods for details on the statistical methods to quantify average signature expression. Data are shown as mean ± standard error of the mean (SEM).
- H Kaplan–Meier analysis representing the probability of metastasis-free survival in breast cancer patients from the metadataset stratified according to high or low glucose NOT YT signature score, which show no differences.
Comment in
- The sweet side of YAP/TAZ.
Santinon G, Enzo E, Dupont S. Santinon G, et al. Cell Cycle. 2015;14(16):2543-4. doi: 10.1080/15384101.2015.1062328. Epub 2015 Jun 26. Cell Cycle. 2015. PMID: 26114316 Free PMC article. No abstract available.
Similar articles
- Metabolic control of YAP and TAZ by the mevalonate pathway.
Sorrentino G, Ruggeri N, Specchia V, Cordenonsi M, Mano M, Dupont S, Manfrin A, Ingallina E, Sommaggio R, Piazza S, Rosato A, Piccolo S, Del Sal G. Sorrentino G, et al. Nat Cell Biol. 2014 Apr;16(4):357-66. doi: 10.1038/ncb2936. Epub 2014 Mar 23. Nat Cell Biol. 2014. PMID: 24658687 - Control of YAP/TAZ Activity by Metabolic and Nutrient-Sensing Pathways.
Santinon G, Pocaterra A, Dupont S. Santinon G, et al. Trends Cell Biol. 2016 Apr;26(4):289-299. doi: 10.1016/j.tcb.2015.11.004. Epub 2015 Dec 30. Trends Cell Biol. 2016. PMID: 26750334 Review. - Regulation of myocardial glucose metabolism by YAP/TAZ signaling.
Kashihara T, Sadoshima J. Kashihara T, et al. J Cardiol. 2024 May;83(5):323-329. doi: 10.1016/j.jjcc.2024.01.002. Epub 2024 Jan 23. J Cardiol. 2024. PMID: 38266816 Review. - A feed forward loop enforces YAP/TAZ signaling during tumorigenesis.
Gill MK, Christova T, Zhang YY, Gregorieff A, Zhang L, Narimatsu M, Song S, Xiong S, Couzens AL, Tong J, Krieger JR, Moran MF, Zlotta AR, van der Kwast TH, Gingras AC, Sicheri F, Wrana JL, Attisano L. Gill MK, et al. Nat Commun. 2018 Aug 29;9(1):3510. doi: 10.1038/s41467-018-05939-2. Nat Commun. 2018. PMID: 30158528 Free PMC article. - A combat with the YAP/TAZ-TEAD oncoproteins for cancer therapy.
Pobbati AV, Hong W. Pobbati AV, et al. Theranostics. 2020 Feb 18;10(8):3622-3635. doi: 10.7150/thno.40889. eCollection 2020. Theranostics. 2020. PMID: 32206112 Free PMC article. Review.
Cited by
- Yap regulates glucose utilization and sustains nucleotide synthesis to enable organ growth.
Cox AG, Tsomides A, Yimlamai D, Hwang KL, Miesfeld J, Galli GG, Fowl BH, Fort M, Ma KY, Sullivan MR, Hosios AM, Snay E, Yuan M, Brown KK, Lien EC, Chhangawala S, Steinhauser ML, Asara JM, Houvras Y, Link B, Vander Heiden MG, Camargo FD, Goessling W. Cox AG, et al. EMBO J. 2018 Nov 15;37(22):e100294. doi: 10.15252/embj.2018100294. Epub 2018 Oct 22. EMBO J. 2018. PMID: 30348863 Free PMC article. - Microarray analysis of breast cancer gene expression profiling in response to 2-deoxyglucose, metformin, and glucose starvation.
Aoun R, El Hadi C, Tahtouh R, El Habre R, Hilal G. Aoun R, et al. Cancer Cell Int. 2022 Mar 19;22(1):123. doi: 10.1186/s12935-022-02542-w. Cancer Cell Int. 2022. PMID: 35305635 Free PMC article. - Nuclear Glycogenolysis Modulates Histone Acetylation in Human Non-Small Cell Lung Cancers.
Sun RC, Dukhande VV, Zhou Z, Young LEA, Emanuelle S, Brainson CF, Gentry MS. Sun RC, et al. Cell Metab. 2019 Nov 5;30(5):903-916.e7. doi: 10.1016/j.cmet.2019.08.014. Epub 2019 Sep 12. Cell Metab. 2019. PMID: 31523006 Free PMC article. - Isoprenylcysteine carboxy methyltransferase (ICMT) is associated with tumor aggressiveness and its expression is controlled by the p53 tumor suppressor.
Borini Etichetti C, Di Benedetto C, Rossi C, Baglioni MV, Bicciato S, Del Sal G, Menacho-Marquez M, Girardini J. Borini Etichetti C, et al. J Biol Chem. 2019 Mar 29;294(13):5060-5073. doi: 10.1074/jbc.RA118.006037. Epub 2019 Jan 17. J Biol Chem. 2019. PMID: 30655292 Free PMC article. - The big picture: exploring the metabolic cross-talk in cancer.
Schulze A, Yuneva M. Schulze A, et al. Dis Model Mech. 2018 Aug 24;11(8):dmm036673. doi: 10.1242/dmm.036673. Dis Model Mech. 2018. PMID: 30154190 Free PMC article.
References
- Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, Solari A, Bobisse S, Rondina MB, Guzzardo V, Parenti AR, Rosato A, Bicciato S, Balmain A, Piccolo S. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. - PubMed
- Alvarez JV, Febbo PG, Ramaswamy S, Loda M, Richardson A, Frank DA. Identification of a genetic signature of activated signal transducer and activator of transcription 3 in human tumors. Cancer Res. 2005;65:5054–5062. - PubMed
- Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–1059. - PubMed
- Atsumi T, Chesney J, Metz C, Leng L, Donnelly S, Makita Z, Mitchell R, Bucala R. High expression of inducible 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (iPFK-2; PFKFB3) in human cancers. Cancer Res. 2002;62:5881–5887. - PubMed
- Azzolin L, Zanconato F, Bresolin S, Forcato M, Basso G, Bicciato S, Cordenonsi M, Piccolo S. Role of TAZ as mediator of Wnt signaling. Cell. 2012;151:1443–1456. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials