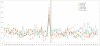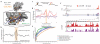Structural imprints in vivo decode RNA regulatory mechanisms - PubMed (original) (raw)
. 2015 Mar 26;519(7544):486-90.
doi: 10.1038/nature14263. Epub 2015 Mar 18.
Affiliations
- PMID: 25799993
- PMCID: PMC4376618
- DOI: 10.1038/nature14263
Structural imprints in vivo decode RNA regulatory mechanisms
Robert C Spitale et al. Nature. 2015.
Erratum in
- Erratum: Structural imprints in vivo decode RNA regulatory mechanisms.
Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, Chang HY. Spitale RC, et al. Nature. 2015 Nov 12;527(7577):264. doi: 10.1038/nature15717. Epub 2015 Sep 30. Nature. 2015. PMID: 26416736 Free PMC article. No abstract available.
Abstract
Visualizing the physical basis for molecular behaviour inside living cells is a great challenge for biology. RNAs are central to biological regulation, and the ability of RNA to adopt specific structures intimately controls every step of the gene expression program. However, our understanding of physiological RNA structures is limited; current in vivo RNA structure profiles include only two of the four nucleotides that make up RNA. Here we present a novel biochemical approach, in vivo click selective 2'-hydroxyl acylation and profiling experiment (icSHAPE), which enables the first global view, to our knowledge, of RNA secondary structures in living cells for all four bases. icSHAPE of the mouse embryonic stem cell transcriptome versus purified RNA folded in vitro shows that the structural dynamics of RNA in the cellular environment distinguish different classes of RNAs and regulatory elements. Structural signatures at translational start sites and ribosome pause sites are conserved from in vitro conditions, suggesting that these RNA elements are programmed by sequence. In contrast, focal structural rearrangements in vivo reveal precise interfaces of RNA with RNA-binding proteins or RNA-modification sites that are consistent with atomic-resolution structural data. Such dynamic structural footprints enable accurate prediction of RNA-protein interactions and N(6)-methyladenosine (m(6)A) modification genome wide. These results open the door for structural genomics of RNA in living cells and reveal key physiological structures controlling gene expression.
Figures
Extended Data Figure 1. Chemical synthesis of NAI-N3
a, Synthetic scheme for NAI-N3. b, 1HMR of Methyl 2-(azidomethyl)nicotinate c, 1HNMR of 2-(azidomethyl)nicotinic acid d, 13CNMR of 2-(azidomethyl)nicotinic acid e, 1HNMR of 2-(azidomethyl)nicotinic acid acyl imidazole f, 13CNMR of 2-(azidomethyl)nicotinic acid acyl imidazole.
Extended Data Figure 2. NAI-N3 is a novel RNA acylation reagent that enables RNA purification
a, Chemical schematic of RNA acylation and copper-free ‘click’ chemistry utilizing NAI-N3 and dibenzocyclooxtyne-biotin conjugate. b, ATP acylation gel shift showing ATP acylation and copper-free ‘click’ chemistry utilizing NAI-N3 and dibenzocyclooxtyne-biotin conjugate.
Extended Data Figure 3. NAI-N3 is a novel RNA acylation reagent accurately reads out RNA structure
a, Comparative denaturing gel of NAI and NAI-N3 RNA acylation. b, Denaturing gel analysis of cDNAs that originate from the biotin-purification protocol (Extended Data Figure 1) c, Secondary structure of the SAM-I Riboswitch with enriched residues highlighted in orange and depleted residues highlighted in blue. d, Denaturing gel analysis of denatured RNA probed with NAI-N3 shows even coverage of 2’-hydroxyl reactivity when RNA is unfolded. e, Protein titration with BSA, demonstrating no difference in the SHAPE pattern as a function of protein concentration.
Extended Data Figure 4. icSHAPE is capable of reproducing RNA acylation profiles obtained by manual RNA modification experiments
icSHAPE (right panels) profiles of ribosomal RNA and compared them to those obtained by manual SHAPE (left panels).
Extended Data Figure 5
RT stops measured by icSHAPE is very well correlated in different library replicates
Extended Data Figure 6. icSHAPE is capable of measuring the RNA structure profiles of thousands of RNAs, simultaneously
a, The RNAs represented in polyA selected RNA, in vivo. b. The RNAs represented in polyA selected RNA, in vitro.
Extended Data Figure 7. Non-AUG start codons are associated with preceding reactivity, and Non-AUG start codons have a different profile, suggesting that RNA accessibility alone is not a sufficient to drive translation
a, icSHAPE profile at AUG start codons, in vivo. b, icSHAPE profile at AUG start codons, in vitro. d, icSHAPE profile at CUG start codons, in vivo. d, icSHAPE profile at CUG start codons, in vitro
Extended Data Figure 8. icSHAPE can be used to predict posttranscriptional regulatory elements
a, icSHAPE profile at Rbfox2 targets, in vivo. b, icSHAPE profile at Rbfox2 targets, in vitro. c, ROC curve of Rbfox2 RNA-protein interactions, predicted using icSHAPE profiles. d, icSHAPE profile at m6A targets, in vivo. The negative control is the set of motif instances that are not m6A modified e, icSHAPE profile at m6A targets, in vitro. f, ROC curve of m6A RNA modification sites, predicted using icSHAPE profiles. g, icSHAPE profile at HuR targets, in vivo. h, icSHAPE profile at HuR targets, in vitro. i, ROC curve of HuR RNA-protein interactions, predicted using icSHAPE profiles.
Extended Data Figure 9. iCLIP analysis of HuR in mESCs
a, global binding preference of the RBP HuR in mESCs as represented by RT stops across the mouse transcriptome (mm9). HuR mainly binds protein coding, processed, and ribosomal RNAs. b, number of unique RNA transcripts bound by HuR. c, HuR RT stops distributed across protein coding transcript functional domains. HuR prefers intronic and 3’UTR regions. d, Metagene analysis of all HuR bindings sites. Each mRNA region (5’UTR, CDS, or 3’UTR) was scaled to a standard unit width RT stop density across all bound protein coding genes was plotted revealing a clear enrichment for 3’UTR regions in mature protein coding transcripts. e, Individual mRNA binding evens of HuR to genes important for mESC biology including Tet1, β-Actin, Elav1 (HuR itself), and Lin28a. Discrete binding sites are observed focused in 3’UTR and intronic regions.
Extended Data Figure 10
m6A-associated RNA structure features are preserved, independent of their position along the RNA transcript
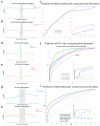
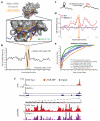
Figure 1. icSHAPE is a Novel and Robust Method for Measuring RNA Structure
a, Chemical scheme for the preparation of acylated RNA, which can be purified by biotin-streptavidin purification. b, Schematic of icSHAPE modification and purification steps to generate a sequencing library. c, Dot blot of biotin-modified RNA from icSHAPE through streptavidin affinity isolation. d, Denaturing gel electrophoresis of icSHAPE on rRNA from mESCs. The corresponding icSHAPE profile, generated from deep sequencing is to the right. e, Pymol representation of rRNA, corresponding to regions of icSHAPE that are more reactive in vivo (PDB3J3D). f, Pymol representation of rRNA, corresponding to regions of icSHAPE that are more reactive in vitro.
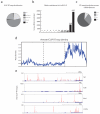
Figure 2. icSHAPE Reveals Unique Structural Profiles for Nucleobase Reactivity and Posttranscriptional Interactions
a, RT stop distribution for the transcriptome, DMSO control, or icSHAPE libraries. b, icSHAPE data track and VTD calculation of the MALAT1 RNA (chr19:5796010-5796081). icSHAPE data are scaled from 0 (no reactivity) to 1 (max reactivity). c, The VTD distribution for icSHAPE libraries. d, GINI index of icSHAPE data in vivo vs. in vitro. e, The distribution of VTD profiles for all hexamer motifs across the transcriptome. Bin locations for several motifs for post-transcriptional regulation are highlighted.
Figure 3. icSHAPE Reveals Structural Profiles Associated with Translation
a, Cartoon representation of ribosomes translating an mRNA. The uORF initiation site is represented by ribosome initiation upstream of the canonical start codon. The canonical start position is demarcated by “AUG”. The N-terminal truncation is represented as a ribosome initiating to the 3’-end of the canonical start “AUG”. b, icSHAPE profile at canonical start codon position. c, icSHAPE profiles at uORF and N-terminal truncation sites. d, Cartoon representation of a paused ribosome and its corresponding A-P-E sites. A: acceptor; P: peptidyl-transferase; E: exit. e, The icSHAPE profile at ribosome pause sites. f, icSHAPE profile at negative control sites for pause sequences. Gray box highlights a region of structural difference upstream of true pause sites vs. controls.
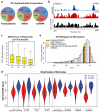
Figure 4. icSHAPE Dynamics Reveals and Predicts Post-transcriptional Interactions
a, Structure of Rbfox1-RNA interaction, highlighting the RNA-protein interface. The RNA is shown with a blue backbone and orange bases; each 2’hydroxyl is green (PDB:2ERR). b, The differential icSHAPE profile at Rbfox2 target mRNAs measured in vivo vs. in vitro maps precisely to the Rbfox binding sites. c, Model of interplay between m6A and RNA structure. d, Differential icSHAPE signal for m6A methylated vs. non-methylated sites with the same underlying sequence motif, both in vivo. icSHAPE signal from unmodified sites are subtracted from m6A-modified sites; asterisks (*) indicate positions with significant differences (p<0.05, FDR<0.05). Data from wild type and Mettl3 KO mESCs are plotted for comparison. e, ROC curve for prediction of m6A sites, incorporating icSHAPE profiles. f, Effect of m6A on RNA structure on Nanog mRNA. Top: location of _Mettl3_-dependent m6A sites (highlight in yellow); m6A-RIP data from Batista et al. Bottom: icSHAPE profile of WT and Mettl3 KO cells.
Figure 5
Comment in
- RNA. Detailed probing of RNA structure in vivo.
Burgess DJ. Burgess DJ. Nat Rev Genet. 2015 May;16(5):255. doi: 10.1038/nrg3939. Epub 2015 Apr 9. Nat Rev Genet. 2015. PMID: 25854184 No abstract available. - Defining Functional Structured RNA inside Living Cells.
Chan D, Spitale RC. Chan D, et al. Biochemistry. 2017 Nov 7;56(44):5847-5848. doi: 10.1021/acs.biochem.7b00816. Biochemistry. 2017. PMID: 29064690 No abstract available.
Similar articles
- Measuring RNA structure transcriptome-wide with icSHAPE.
Chan D, Feng C, Spitale RC. Chan D, et al. Methods. 2017 May 1;120:85-90. doi: 10.1016/j.ymeth.2017.02.010. Epub 2017 Mar 20. Methods. 2017. PMID: 28336307 Free PMC article. Review. - Transcriptome-wide interrogation of RNA secondary structure in living cells with icSHAPE.
Flynn RA, Zhang QC, Spitale RC, Lee B, Mumbach MR, Chang HY. Flynn RA, et al. Nat Protoc. 2016 Feb;11(2):273-90. doi: 10.1038/nprot.2016.011. Epub 2016 Jan 14. Nat Protoc. 2016. PMID: 26766114 Free PMC article. - RNA structure maps across mammalian cellular compartments.
Sun L, Fazal FM, Li P, Broughton JP, Lee B, Tang L, Huang W, Kool ET, Chang HY, Zhang QC. Sun L, et al. Nat Struct Mol Biol. 2019 Apr;26(4):322-330. doi: 10.1038/s41594-019-0200-7. Epub 2019 Mar 18. Nat Struct Mol Biol. 2019. PMID: 30886404 Free PMC article. - In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features.
Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. Ding Y, et al. Nature. 2014 Jan 30;505(7485):696-700. doi: 10.1038/nature12756. Epub 2013 Nov 24. Nature. 2014. PMID: 24270811 - RNA binding protein/RNA element interactions and the control of translation.
Pichon X, Wilson LA, Stoneley M, Bastide A, King HA, Somers J, Willis AE. Pichon X, et al. Curr Protein Pept Sci. 2012 Jun;13(4):294-304. doi: 10.2174/138920312801619475. Curr Protein Pept Sci. 2012. PMID: 22708490 Free PMC article. Review.
Cited by
- diffBUM-HMM: a robust statistical modeling approach for detecting RNA flexibility changes in high-throughput structure probing data.
Marangio P, Law KYT, Sanguinetti G, Granneman S. Marangio P, et al. Genome Biol. 2021 May 27;22(1):165. doi: 10.1186/s13059-021-02379-y. Genome Biol. 2021. PMID: 34044851 Free PMC article. - RNA structure determination: From 2D to 3D.
Deng J, Fang X, Huang L, Li S, Xu L, Ye K, Zhang J, Zhang K, Zhang QC. Deng J, et al. Fundam Res. 2023 Jun 12;3(5):727-737. doi: 10.1016/j.fmre.2023.06.001. eCollection 2023 Sep. Fundam Res. 2023. PMID: 38933295 Free PMC article. Review. - RNA. Detailed probing of RNA structure in vivo.
Burgess DJ. Burgess DJ. Nat Rev Genet. 2015 May;16(5):255. doi: 10.1038/nrg3939. Epub 2015 Apr 9. Nat Rev Genet. 2015. PMID: 25854184 No abstract available. - N(6)-Methyladenosine: a conformational marker that regulates the substrate specificity of human demethylases FTO and ALKBH5.
Zou S, Toh JD, Wong KH, Gao YG, Hong W, Woon EC. Zou S, et al. Sci Rep. 2016 May 9;6:25677. doi: 10.1038/srep25677. Sci Rep. 2016. PMID: 27156733 Free PMC article. - m(6)A-LAIC-seq reveals the census and complexity of the m(6)A epitranscriptome.
Molinie B, Wang J, Lim KS, Hillebrand R, Lu ZX, Van Wittenberghe N, Howard BD, Daneshvar K, Mullen AC, Dedon P, Xing Y, Giallourakis CC. Molinie B, et al. Nat Methods. 2016 Aug;13(8):692-8. doi: 10.1038/nmeth.3898. Epub 2016 Jul 4. Nat Methods. 2016. PMID: 27376769 Free PMC article.
References
- Ding Y, et al. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi:10.1038/nature12756. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 CA009302/CA/NCI NIH HHS/United States
- R01HG004361/HG/NHGRI NIH HHS/United States
- R01 HG004361/HG/NHGRI NIH HHS/United States
- F30CA189514/CA/NCI NIH HHS/United States
- P50 HG007735/HG/NHGRI NIH HHS/United States
- P50HG007735/HG/NHGRI NIH HHS/United States
- Howard Hughes Medical Institute/United States
- R01 GM068122/GM/NIGMS NIH HHS/United States
- T32AR007422/AR/NIAMS NIH HHS/United States
- F30 CA189514/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases