Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology - PubMed (original) (raw)
doi: 10.1016/j.jmoldx.2014.12.006. Epub 2015 Mar 20.
Talia N Mitchell 1, Ahmet Zehir 1, Ronak H Shah 1, Ryma Benayed 1, Aijazuddin Syed 1, Raghu Chandramohan 1, Zhen Yu Liu 1, Helen H Won 1, Sasinya N Scott 1, A Rose Brannon 1, Catherine O'Reilly 1, Justyna Sadowska 1, Jacklyn Casanova 1, Angela Yannes 1, Jaclyn F Hechtman 1, Jinjuan Yao 1, Wei Song 1, Dara S Ross 1, Alifya Oultache 1, Snjezana Dogan 1, Laetitia Borsu 1, Meera Hameed 1, Khedoudja Nafa 1, Maria E Arcila 1, Marc Ladanyi 2, Michael F Berger 3
Affiliations
- PMID: 25801821
- PMCID: PMC5808190
- DOI: 10.1016/j.jmoldx.2014.12.006
Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology
Donavan T Cheng et al. J Mol Diagn. 2015 May.
Abstract
The identification of specific genetic alterations as key oncogenic drivers and the development of targeted therapies are together transforming clinical oncology and creating a pressing need for increased breadth and throughput of clinical genotyping. Next-generation sequencing assays allow the efficient and unbiased detection of clinically actionable mutations. To enable precision oncology in patients with solid tumors, we developed Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT), a hybridization capture-based next-generation sequencing assay for targeted deep sequencing of all exons and selected introns of 341 key cancer genes in formalin-fixed, paraffin-embedded tumors. Barcoded libraries from patient-matched tumor and normal samples were captured, sequenced, and subjected to a custom analysis pipeline to identify somatic mutations. Sensitivity, specificity, reproducibility of MSK-IMPACT were assessed through extensive analytical validation. We tested 284 tumor samples with previously known point mutations and insertions/deletions in 47 exons of 19 cancer genes. All known variants were accurately detected, and there was high reproducibility of inter- and intrarun replicates. The detection limit for low-frequency variants was approximately 2% for hotspot mutations and 5% for nonhotspot mutations. Copy number alterations and structural rearrangements were also reliably detected. MSK-IMPACT profiles oncogenic DNA alterations in clinical solid tumor samples with high accuracy and sensitivity. Paired analysis of tumors and patient-matched normal samples enables unambiguous detection of somatic mutations to guide treatment decisions.
Copyright © 2015 American Society for Investigative Pathology and the Association for Molecular Pathology. Published by Elsevier Inc. All rights reserved.
Figures
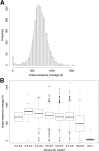
Figure 1
Uniformity of sequence coverage. A: Distribution of mean coverage values across all canonical exons in 341 genes targeted by MSK-IMPACT. Coverage values were computed using a panel of 10 diploid, normal formalin-fixed, paraffin-embedded samples, each run in duplicate. B: Coverage for canonical exons binned by percent GC content.
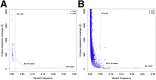
Figure 2
Selection of variant calling filters. Coverage and variant frequency values are plotted for false-positive variant calls generated by comparing experimental replicates of normal formalin-fixed, paraffin-embedded samples against each other. A: First-tier false-positive calls occurring in hotspot regions. B: Second-tier false-positive calls occurring outside hotspot regions. Dotted lines indicate decision boundaries for rejecting false positives based on thresholds on coverage (DP ≥20×), number of mutant reads (AD ≥8 reads for first-tier events, AD ≥10 reads for second-tier events), and variant frequency (VF ≥2% for first-tier events, VF ≥5% for second-tier events). AD, number of mutant reads; DP, coverage depth; Indel, insertion/deletion; SNV, single nucleotide variation; VF, variant frequency.
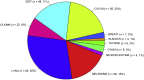
Figure 3
Distribution of tumor types among the 284 unique samples profiled for the clinical validation study.
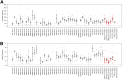
Figure 4
Range of coverage depth values (A) and variant frequencies (B) for known variants detected in 47 exons of 19 genes tested. Gray indicates SNVs, red, indels. Bars indicate mean values, whiskers indicate SEM. indel, insertion/deletion; SNV, single nucleotide variation.
Figure 5
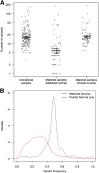
Variant calling for unmatched tumors. A: Number of variants called on: i) 209 samples for which a matched normal was unavailable, 75 samples where matched normal samples were available; and ii) variant calling was performed against the matched normal; or iii) variant calling was deliberately performed using a generic pooled normal. B: Distribution of variant frequencies for somatic mutations (red line) and additional private germline mutations (black line) identified when a generic pooled normal is used. Bars indicate mean values, whiskers indicate SEM.
Figure 6
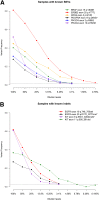
Variant frequencies of known SNV (A) and indel (B) calls tracked across successive serial dilutions. indel, insertion/deletion; SNV, single nucleotide variation.
Figure 7
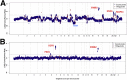
MSK-IMPACT reveals copy number alterations. Log-ratios comparing tumor versus normal coverage values are calculated across all targeted regions for samples containing ERBB2 amplifications: breast cancer sample with ERBB2, GNAS, MAPK1 amplifications, and ATM loss (A), gastric cancer sample with EGFR, ERBB2, and PIM1 amplifications (B).
Figure 8
MSK-IMPACT reveals structural rearrangements. Integrated Genomics Viewer screenshot of _EML4_-ALK translocation in a lung adenocarcinoma sample known to be positive for this translocation.
Similar articles
- Comprehensive detection of germline variants by MSK-IMPACT, a clinical diagnostic platform for solid tumor molecular oncology and concurrent cancer predisposition testing.
Cheng DT, Prasad M, Chekaluk Y, Benayed R, Sadowska J, Zehir A, Syed A, Wang YE, Somar J, Li Y, Yelskaya Z, Wong D, Robson ME, Offit K, Berger MF, Nafa K, Ladanyi M, Zhang L. Cheng DT, et al. BMC Med Genomics. 2017 May 19;10(1):33. doi: 10.1186/s12920-017-0271-4. BMC Med Genomics. 2017. PMID: 28526081 Free PMC article. - TumorNext: A comprehensive tumor profiling assay that incorporates high resolution copy number analysis and germline status to improve testing accuracy.
Gray PN, Vuong H, Tsai P, Lu HM, Mu W, Hsuan V, Hoo J, Shah S, Uyeda L, Fox S, Patel H, Janicek M, Brown S, Dobrea L, Wagman L, Plimack E, Mehra R, Golemis EA, Bilusic M, Zibelman M, Elliott A. Gray PN, et al. Oncotarget. 2016 Oct 18;7(42):68206-68228. doi: 10.18632/oncotarget.11910. Oncotarget. 2016. PMID: 27626691 Free PMC article. - Validation of OncoPanel: A Targeted Next-Generation Sequencing Assay for the Detection of Somatic Variants in Cancer.
Garcia EP, Minkovsky A, Jia Y, Ducar MD, Shivdasani P, Gong X, Ligon AH, Sholl LM, Kuo FC, MacConaill LE, Lindeman NI, Dong F. Garcia EP, et al. Arch Pathol Lab Med. 2017 Jun;141(6):751-758. doi: 10.5858/arpa.2016-0527-OA. Epub 2017 Mar 3. Arch Pathol Lab Med. 2017. PMID: 28557599 - Next Generation Sequencing for the Detection of Actionable Mutations in Solid and Liquid Tumors.
Fox AJ, Hiemenz MC, Lieberman DB, Sukhadia S, Li B, Grubb J, Candrea P, Ganapathy K, Zhao J, Roth D, Alley E, Loren A, Morrissette JJ. Fox AJ, et al. J Vis Exp. 2016 Sep 20;(115):52758. doi: 10.3791/52758. J Vis Exp. 2016. PMID: 27684276 Free PMC article. Review. - The possibility of clinical sequencing in the management of cancer.
Kou T, Kanai M, Matsumoto S, Okuno Y, Muto M. Kou T, et al. Jpn J Clin Oncol. 2016 May;46(5):399-406. doi: 10.1093/jjco/hyw018. Epub 2016 Feb 24. Jpn J Clin Oncol. 2016. PMID: 26917600 Review.
Cited by
- A Phase Ib Study of Alpelisib (BYL719), a PI3Kα-Specific Inhibitor, with Letrozole in ER+/HER2- Metastatic Breast Cancer.
Mayer IA, Abramson VG, Formisano L, Balko JM, Estrada MV, Sanders ME, Juric D, Solit D, Berger MF, Won HH, Li Y, Cantley LC, Winer E, Arteaga CL. Mayer IA, et al. Clin Cancer Res. 2017 Jan 1;23(1):26-34. doi: 10.1158/1078-0432.CCR-16-0134. Epub 2016 Apr 28. Clin Cancer Res. 2017. PMID: 27126994 Free PMC article. Clinical Trial. - A novel tumor mutational burden estimation model as a predictive and prognostic biomarker in NSCLC patients.
Tian Y, Xu J, Chu Q, Duan J, Zhang J, Bai H, Yang Z, Fang W, Cai L, Wan R, Fei K, He J, Gao S, Zhang L, Wang Z, Wang J. Tian Y, et al. BMC Med. 2020 Aug 26;18(1):232. doi: 10.1186/s12916-020-01694-8. BMC Med. 2020. PMID: 32843031 Free PMC article. - Sentinel lymph node mapping in patients with endometrial hyperplasia: A practice to preserve or abandon?
Mueller JJ, Rios-Doria E, Park KJ, Broach VA, Alektiar KM, Jewell EL, Zivanovic O, Sonoda Y, Abu-Rustum NR, Leitao MM Jr, Gardner GJ. Mueller JJ, et al. Gynecol Oncol. 2023 Jan;168:1-7. doi: 10.1016/j.ygyno.2022.10.017. Epub 2022 Nov 2. Gynecol Oncol. 2023. PMID: 36334496 Free PMC article. - YAP localization mediates mechanical adaptation of human cancer cells during extravasation in vivo.
So WY, Wong CS, Azubuike UF, Paul CD, Sangsari PR, Gordon PB, Gong H, Maity TK, Lim P, Yang Z, Haryanto CA, Batchelor E, Jenkins LM, Morgan NY, Tanner K. So WY, et al. bioRxiv [Preprint]. 2023 Nov 16:2023.11.14.567015. doi: 10.1101/2023.11.14.567015. bioRxiv. 2023. PMID: 38076880 Free PMC article. Preprint. - Folate receptor alpha expression in low-grade serous ovarian cancer: Exploring new therapeutic possibilities.
Manning-Geist BL, Sullivan MW, Zhou Q, Iasonos A, Selenica P, Stallworth C, Liu YL, Long Roche K, Gordhandas S, Aghajanian C, Chi D, O'Cearbhaill R, Grisham RN, Chui MH. Manning-Geist BL, et al. Gynecol Oncol. 2024 Sep;188:52-57. doi: 10.1016/j.ygyno.2024.06.008. Epub 2024 Jun 27. Gynecol Oncol. 2024. PMID: 38941962
References
- Garraway L.A. Genomics-driven oncology: framework for an emerging paradigm. J Clin Oncol. 2013;31:1806–1814. - PubMed
- Taylor B.S., Ladanyi M. Clinical cancer genomics: how soon is now? J Pathol. 2011;223:318–326. - PubMed
- Romano E., Schwartz G.K., Chapman P.B., Wolchock J.D., Carvajal R.D. Treatment implications of the emerging molecular classification system for melanoma. Lancet Oncol. 2011;12:913–922. - PubMed
- Chapman P.B., Hauschild A., Robert C., Haanen J.B., Ascierto P., Larkin J., Dummer R., Garbe C., Testori A., Maio M., Hogg D., Lorigan P., Lebbe C., Jouary T., Schadendorf D., Ribas A., O'Day S.J., Sosman J.A., Kirkwood J.M., Eggermont A.M., Dreno B., Nolop K., Li J., Nelson B., Hou J., Lee R.J., Flaherty K.T., McArthur G.A. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources