Unravelling the hidden ancestry of American admixed populations - PubMed (original) (raw)
Unravelling the hidden ancestry of American admixed populations
Francesco Montinaro et al. Nat Commun. 2015.
Abstract
The movement of people into the Americas has brought different populations into contact, and contemporary American genomes are the product of a range of complex admixture events. Here we apply a haplotype-based ancestry identification approach to a large set of genome-wide SNP data from a variety of American, European and African populations to determine the contributions of different ancestral populations to the Americas. Our results provide a fine-scale characterization of the source populations, identify a series of novel, previously unreported contributions from Africa and Europe and highlight geohistorical structure in the ancestry of American admixed populations.
Figures
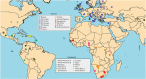
Figure 1. Approximate geographic sampling location of donor and recipient populations analysed.
Colours refer to the 13 groups as described in Fig. 2 and Supplementary Table 2. Circles and diamond refer, respectively, to donors and recipients.
Figure 2. fineSTRUCTURE clustering of the analysed individuals.
Tree of the analysed individuals pooled in 78 clusters as inferred by fineSTRUCTURE. Colours follow macro-area affiliations as in Fig. 1.
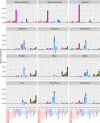
Figure 3. Contribution of the most by informative 23 clusters inferred by fineSTRUCTURE to the analysed recipient populations.
Contribution of the most informative 23 clusters to the American and Caribbean populations estimated using the non-negative least square approach. Standard error based on jack-knife resampling (22 replicates) is reported.
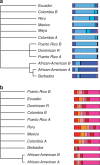
Figure 4. Hierarchical consensus trees of the continental components for American and Caribbean populations.
Consensus tree using Hierarchical clustering for (a) European component; (b) African component. Bar plots at the tips of the trees indicate the relative ancestry composition of the analysed population; colours refer to the 13 groups in Fig. 1. Only branches supported by more than 80% of the 1,000 trees built by bootstrap described in Methods are retained.
Similar articles
- Ancestral components of admixed genomes in a Mexican cohort.
Johnson NA, Coram MA, Shriver MD, Romieu I, Barsh GS, London SJ, Tang H. Johnson NA, et al. PLoS Genet. 2011 Dec;7(12):e1002410. doi: 10.1371/journal.pgen.1002410. Epub 2011 Dec 15. PLoS Genet. 2011. PMID: 22194699 Free PMC article. - The Genomic Impact of European Colonization of the Americas.
Ongaro L, Scliar MO, Flores R, Raveane A, Marnetto D, Sarno S, Gnecchi-Ruscone GA, Alarcón-Riquelme ME, Patin E, Wangkumhang P, Hellenthal G, Gonzalez-Santos M, King RJ, Kouvatsi A, Balanovsky O, Balanovska E, Atramentova L, Turdikulova S, Mastana S, Marjanovic D, Mulahasanovic L, Leskovac A, Lima-Costa MF, Pereira AC, Barreto ML, Horta BL, Mabunda N, May CA, Moreno-Estrada A, Achilli A, Olivieri A, Semino O, Tambets K, Kivisild T, Luiselli D, Torroni A, Capelli C, Tarazona-Santos E, Metspalu M, Pagani L, Montinaro F. Ongaro L, et al. Curr Biol. 2019 Dec 2;29(23):3974-3986.e4. doi: 10.1016/j.cub.2019.09.076. Epub 2019 Nov 14. Curr Biol. 2019. PMID: 31735679 - Development of a panel of genome-wide ancestry informative markers to study admixture throughout the Americas.
Galanter JM, Fernandez-Lopez JC, Gignoux CR, Barnholtz-Sloan J, Fernandez-Rozadilla C, Via M, Hidalgo-Miranda A, Contreras AV, Figueroa LU, Raska P, Jimenez-Sanchez G, Zolezzi IS, Torres M, Ponte CR, Ruiz Y, Salas A, Nguyen E, Eng C, Borjas L, Zabala W, Barreto G, González FR, Ibarra A, Taboada P, Porras L, Moreno F, Bigham A, Gutierrez G, Brutsaert T, León-Velarde F, Moore LG, Vargas E, Cruz M, Escobedo J, Rodriguez-Santana J, Rodriguez-Cintrón W, Chapela R, Ford JG, Bustamante C, Seminara D, Shriver M, Ziv E, Burchard EG, Haile R, Parra E, Carracedo A; LACE Consortium. Galanter JM, et al. PLoS Genet. 2012;8(3):e1002554. doi: 10.1371/journal.pgen.1002554. Epub 2012 Mar 8. PLoS Genet. 2012. PMID: 22412386 Free PMC article. - Mapping of disease-associated variants in admixed populations.
Shriner D, Adeyemo A, Ramos E, Chen G, Rotimi CN. Shriner D, et al. Genome Biol. 2011;12(5):223. doi: 10.1186/gb-2011-12-5-223. Epub 2011 May 30. Genome Biol. 2011. PMID: 21635713 Free PMC article. Review. - The Genetic Diversity of the Americas.
Adhikari K, Chacón-Duque JC, Mendoza-Revilla J, Fuentes-Guajardo M, Ruiz-Linares A. Adhikari K, et al. Annu Rev Genomics Hum Genet. 2017 Aug 31;18:277-296. doi: 10.1146/annurev-genom-083115-022331. Annu Rev Genomics Hum Genet. 2017. PMID: 28859572 Review.
Cited by
- AFA: Ancestry-specific allele frequency estimation in admixed populations: The Hispanic Community Health Study/Study of Latinos.
Granot-Hershkovitz E, Sun Q, Argos M, Zhou H, Lin X, Browning SR, Sofer T. Granot-Hershkovitz E, et al. HGG Adv. 2022 Feb 24;3(2):100096. doi: 10.1016/j.xhgg.2022.100096. eCollection 2022 Apr 14. HGG Adv. 2022. PMID: 35300209 Free PMC article. - A framework for research into continental ancestry groups of the UK Biobank.
Constantinescu AE, Mitchell RE, Zheng J, Bull CJ, Timpson NJ, Amulic B, Vincent EE, Hughes DA. Constantinescu AE, et al. Hum Genomics. 2022 Jan 29;16(1):3. doi: 10.1186/s40246-022-00380-5. Hum Genomics. 2022. PMID: 35093177 Free PMC article. - Multiple Sources of Introduction of North American Arabidopsis thaliana from across Eurasia.
Shirsekar G, Devos J, Latorre SM, Blaha A, Queiroz Dias M, González Hernando A, Lundberg DS, Burbano HA, Fenster CB, Weigel D. Shirsekar G, et al. Mol Biol Evol. 2021 Dec 9;38(12):5328-5344. doi: 10.1093/molbev/msab268. Mol Biol Evol. 2021. PMID: 34499163 Free PMC article. - Evaluation of loci to predict ear morphology using two SNaPshot assays.
Noreen S, Ballard D, Mehmood T, Khan A, Khalid T, Rakha A. Noreen S, et al. Forensic Sci Med Pathol. 2023 Sep;19(3):335-356. doi: 10.1007/s12024-022-00545-7. Epub 2022 Nov 19. Forensic Sci Med Pathol. 2023. PMID: 36401782 Free PMC article. - The Genetic Ancestry of Modern Indus Valley Populations from Northwest India.
Pathak AK, Kadian A, Kushniarevich A, Montinaro F, Mondal M, Ongaro L, Singh M, Kumar P, Rai N, Parik J, Metspalu E, Rootsi S, Pagani L, Kivisild T, Metspalu M, Chaubey G, Villems R. Pathak AK, et al. Am J Hum Genet. 2018 Dec 6;103(6):918-929. doi: 10.1016/j.ajhg.2018.10.022. Am J Hum Genet. 2018. PMID: 30526867 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
- 098387/WT_/Wellcome Trust/United Kingdom
- 098387/Z/12/Z/WT_/Wellcome Trust/United Kingdom
- 098386/Z/12/Z/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 098386/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical