Global genetic analysis in mice unveils central role for cilia in congenital heart disease - PubMed (original) (raw)
. 2015 May 28;521(7553):520-4.
doi: 10.1038/nature14269. Epub 2015 Mar 25.
Nikolai T Klena 1, George C Gabriel 1, Xiaoqin Liu 1, Andrew J Kim 1, Kristi Lemke 1, Yu Chen 1, Bishwanath Chatterjee 1, William Devine 2, Rama Rao Damerla 1, Chienfu Chang 1, Hisato Yagi 1, Jovenal T San Agustin 3, Mohamed Thahir 4, Shane Anderton 1, Caroline Lawhead 1, Anita Vescovi 1, Herbert Pratt 5, Judy Morgan 5, Leslie Haynes 5, Cynthia L Smith 5, Janan T Eppig 5, Laura Reinholdt 5, Richard Francis 1, Linda Leatherbury 6, Madhavi K Ganapathiraju 4, Kimimasa Tobita 1, Gregory J Pazour 3, Cecilia W Lo 1
Affiliations
- PMID: 25807483
- PMCID: PMC4617540
- DOI: 10.1038/nature14269
Global genetic analysis in mice unveils central role for cilia in congenital heart disease
You Li et al. Nature. 2015.
Abstract
Congenital heart disease (CHD) is the most prevalent birth defect, affecting nearly 1% of live births; the incidence of CHD is up to tenfold higher in human fetuses. A genetic contribution is strongly suggested by the association of CHD with chromosome abnormalities and high recurrence risk. Here we report findings from a recessive forward genetic screen in fetal mice, showing that cilia and cilia-transduced cell signalling have important roles in the pathogenesis of CHD. The cilium is an evolutionarily conserved organelle projecting from the cell surface with essential roles in diverse cellular processes. Using echocardiography, we ultrasound scanned 87,355 chemically mutagenized C57BL/6J fetal mice and recovered 218 CHD mouse models. Whole-exome sequencing identified 91 recessive CHD mutations in 61 genes. This included 34 cilia-related genes, 16 genes involved in cilia-transduced cell signalling, and 10 genes regulating vesicular trafficking, a pathway important for ciliogenesis and cell signalling. Surprisingly, many CHD genes encoded interacting proteins, suggesting that an interactome protein network may provide a larger genomic context for CHD pathogenesis. These findings provide novel insights into the potential Mendelian genetic contribution to CHD in the fetal population, a segment of the human population not well studied. We note that the pathways identified show overlap with CHD candidate genes recovered in CHD patients, suggesting that they may have relevance to the more complex genetics of CHD overall. These CHD mouse models and >8,000 incidental mutations have been sperm archived, creating a rich public resource for human disease modelling.
Figures
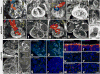
Figure 1. Ultrasound diagnoses of CHD and cilia defects in CHD mutants
Vevo 2100 color flow imaging showed criss-crossing of blood flow indicating normal aorta (Ao) and pulmonary artery (PA) alignment (a, Supplemental Movie S1), confirmed by histopathology (b). E16.5 mutant (line b2b327) exhibited blood flow pattern indicating single great artery (PA) and ventricular septal defect (VSD) (c, Supplemental Movie S2), suggesting aortic atresia with VSD, confirmed by histopathology (d). Color flow imaging of E15.5 mutant (line b2b2025) with heterotaxy (stomach on right; Supplemental Movie S3c) had side-by-side Ao/PA with Ao emerging from right ventricle (RV), indicating DORV/VSD (e,f,Supplemental Movie S3a), and presence of AVSD (g,h,Supplemental Movie S3b,S3c). Histopathology also showed bicuspid aortic valve (BAV,i), interrupted aortic arch (IAA,j), and common AV valve (k). (l-n). Cc2d2a mutant exhibits dextrocardia with ventricular inversion (dextroversion) (m), and AVSD (l) with malformed AV cushions (n), but normal outflow cushions. (o-x). Confocal imaging of E12.5 Cc2d2a mutant (m/m) vs. wildtype (+/+) embryo sections showed no cilia in AV cushion (o,p), but normal ciliation in outflow cushion (q,r). Fewer and shorter cilia were observed in other mutant embryo tissues (s-x). Red:acetylated tubulin;green:IFT88.
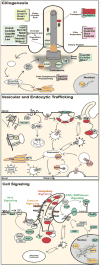
Figure 2. Congenital heart disease genes recovered from mouse mutagenesis screen
Diagrams illustrate biological context of CHD gene function (color-highlighting indicate CHD genes recovered; asterisk denote CHD genes recovered from previous screen). For clarity, ciliome genes (Dctn5,Fuz) with endocytic function and/or involved in cilia-transduced signaling were not shown in the ciliogenesis panel.
Figure 3. Interactome network of CHD genes with known and predicted interactors
An interactome network was constructed (a) comprising the 61 CHD genes (magenta squares) and their known (blue edges) and predicted (red edges) interactions. The CHD interactome yielded significant enrichment of GO terms related to developmental processes. Circle size proportional to number of genes associated with GO terms.
Similar articles
- The Role of Cilia and the Complex Genetics of Congenital Heart Disease.
Gabriel GC, Ganapathiraju M, Lo CW. Gabriel GC, et al. Annu Rev Genomics Hum Genet. 2024 Aug;25(1):309-327. doi: 10.1146/annurev-genom-121222-105345. Epub 2024 Aug 6. Annu Rev Genomics Hum Genet. 2024. PMID: 38724024 Review. - Role of cilia in the pathogenesis of congenital heart disease.
Gabriel GC, Young CB, Lo CW. Gabriel GC, et al. Semin Cell Dev Biol. 2021 Feb;110:2-10. doi: 10.1016/j.semcdb.2020.04.017. Epub 2020 May 14. Semin Cell Dev Biol. 2021. PMID: 32418658 Free PMC article. Review. - Role of cilia in structural birth defects: insights from ciliopathy mutant mouse models.
Rao Damerla R, Gabriel GC, Li Y, Klena NT, Liu X, Chen Y, Cui C, Pazour GJ, Lo CW. Rao Damerla R, et al. Birth Defects Res C Embryo Today. 2014 Jun;102(2):115-25. doi: 10.1002/bdrc.21067. Birth Defects Res C Embryo Today. 2014. PMID: 24975753 Review. - Interrogating congenital heart defects with noninvasive fetal echocardiography in a mouse forward genetic screen.
Liu X, Francis R, Kim AJ, Ramirez R, Chen G, Subramanian R, Anderton S, Kim Y, Wong L, Morgan J, Pratt HC, Reinholdt L, Devine W, Leatherbury L, Tobita K, Lo CW. Liu X, et al. Circ Cardiovasc Imaging. 2014 Jan;7(1):31-42. doi: 10.1161/CIRCIMAGING.113.000451. Epub 2013 Dec 6. Circ Cardiovasc Imaging. 2014. PMID: 24319090 Free PMC article. - Cilia and Ciliopathies in Congenital Heart Disease.
Klena NT, Gibbs BC, Lo CW. Klena NT, et al. Cold Spring Harb Perspect Biol. 2017 Aug 1;9(8):a028266. doi: 10.1101/cshperspect.a028266. Cold Spring Harb Perspect Biol. 2017. PMID: 28159874 Free PMC article. Review.
Cited by
- Genome-Wide Association Study of Down Syndrome-Associated Atrioventricular Septal Defects.
Ramachandran D, Zeng Z, Locke AE, Mulle JG, Bean LJ, Rosser TC, Dooley KJ, Cua CL, Capone GT, Reeves RH, Maslen CL, Cutler DJ, Feingold E, Sherman SL, Zwick ME. Ramachandran D, et al. G3 (Bethesda). 2015 Jul 20;5(10):1961-71. doi: 10.1534/g3.115.019943. G3 (Bethesda). 2015. PMID: 26194203 Free PMC article. - Aggrecan in Cardiovascular Development and Disease.
Koch CD, Lee CM, Apte SS. Koch CD, et al. J Histochem Cytochem. 2020 Nov;68(11):777-795. doi: 10.1369/0022155420952902. Epub 2020 Sep 1. J Histochem Cytochem. 2020. PMID: 32870742 Free PMC article. Review. - A CEP104-CSPP1 Complex Is Required for Formation of Primary Cilia Competent in Hedgehog Signaling.
Frikstad KM, Molinari E, Thoresen M, Ramsbottom SA, Hughes F, Letteboer SJF, Gilani S, Schink KO, Stokke T, Geimer S, Pedersen LB, Giles RH, Akhmanova A, Roepman R, Sayer JA, Patzke S. Frikstad KM, et al. Cell Rep. 2019 Aug 13;28(7):1907-1922.e6. doi: 10.1016/j.celrep.2019.07.025. Cell Rep. 2019. PMID: 31412255 Free PMC article. - Rare Copy Number Variants Identify Novel Genes in Sporadic Total Anomalous Pulmonary Vein Connection.
Shi X, Cheng L, Jiao X, Chen B, Li Z, Liang Y, Liu W, Wang J, Liu G, Xu Y, Sun J, Fu Q, Lu Y, Chen S. Shi X, et al. Front Genet. 2018 Nov 23;9:559. doi: 10.3389/fgene.2018.00559. eCollection 2018. Front Genet. 2018. PMID: 30532766 Free PMC article. - Genetics of Congenital Heart Disease.
Williams K, Carson J, Lo C. Williams K, et al. Biomolecules. 2019 Dec 16;9(12):879. doi: 10.3390/biom9120879. Biomolecules. 2019. PMID: 31888141 Free PMC article. Review.
References
- Hoffman JI. Incidence of congenital heart disease: II. Prenatal incidence. Pediatric cardiology. 1995;16:155–165. - PubMed
Publication types
MeSH terms
Grants and funding
- HG000330/HG/NHGRI NIH HHS/United States
- U01 HL098188/HL/NHLBI NIH HHS/United States
- U01HL098188/HL/NHLBI NIH HHS/United States
- P41 HG000330/HG/NHGRI NIH HHS/United States
- U41 HG000330/HG/NHGRI NIH HHS/United States
- U01 HL098180/HL/NHLBI NIH HHS/United States
- R01 MH094564/MH/NIMH NIH HHS/United States
- U01HL098180/HL/NHLBI NIH HHS/United States
- R01 GM060992/GM/NIGMS NIH HHS/United States
- R01MH094564/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials