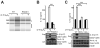RNase L activates the NLRP3 inflammasome during viral infections - PubMed (original) (raw)
RNase L activates the NLRP3 inflammasome during viral infections
Arindam Chakrabarti et al. Cell Host Microbe. 2015.
Abstract
The NLRP3 inflammasome assembles in response to danger signals, triggering self-cleavage of procaspase-1 and production of the proinflammatory cytokine IL-1β. Although virus infection activates the NLRP3 inflammasome, the underlying events remain incompletely understood. We report that virus activation of the NLRP3 inflammasome involves the 2',5'-oligoadenylate (2-5A) synthetase(OAS)/RNase L system, a component of the interferon-induced antiviral response that senses double-stranded RNA and activates endoribonuclease RNase L to cleave viral and cellular RNAs. The absence of RNase L reduces IL-1β production in influenza A virus-infected mice. RNA cleavage products generated by RNase L enhance IL-1β production but require the presence of 2',3'-cyclic phosphorylated termini characteristic of RNase L activity. Additionally, these cleavage products stimulate NLRP3 complex formation with the DExD/H-box helicase, DHX33, and mitochondrial adaptor protein, MAVS, which are each required for effective NLRP3 inflammasome activation. Thus, RNA cleavage events catalyzed by RNase L are required for optimal inflammasome activation during viral infections.
Copyright © 2015 Elsevier Inc. All rights reserved.
Conflict of interest statement
CONFLICT OF INTEREST STATEMENT
Co-author Luigi Franchi is currently an employee of Lycera, a biotechnology company that develops drugs for inflammation. The other co-authors do not have a conflict of interest.
Figures
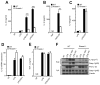
Figure 1. RNase L deficiency in mice enhances the lethality of IAV while decreasing the induction of IL-1β
(A) Survival of wt and _Rnasel_−/− mice infected i.n. with IAV. (B) Mean body weights corresponding to panel (A). (C,D) IL-1β and (E,F) TNF-α levels determined by ELISAs at 2 days post-infection (dpi) with IAV in (C,E) lung homogenates and (D,F) BALF. (G) Viral titers from lungs harvested at 7 dpi, determined by plaque assays. horizontal lines, mean ± standard deviation (SD). Significance was determined by (A,B) Kaplan-Meier analysis or (C–G) two-tailed Student’s t tests. *** p<0.001; **p< 0.01; *p<0.05; ns, not significant. Numbers of mice used: (A,B) n= 14; (C,E) n=8; (D) n=11; (F) n=9; and (G) n=8. See also Figure S1.
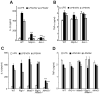
Figure 2. Inflammasome activation in response to viral infections is reduced in the absence of RNase L
IL-1β levels by ELISAs from (A) wt and Rnasel−/− BMDC primed with LPS for 6 hrs and infected with VSV for 12 hrs or IAV for 24 hrs, (B) wt and Rnasel−/− BMDC primed with TNFα for 6 hrs and infected with VSV for 12 hrs and (C) BMDC primed with LPS for 6 hrs and treated with extracellular ATP (0.5 mM) for 1 hr. (D) IL-1β mRNA levels by qRT-PCR and (E) TNFα levels by ELISAs from LPS primed wt and Rnasel−/− BMDCs without or with VSV infection. (F) Cleavage of caspase-1 and pro-IL-1β in LPS primed wt and Rnasel−/− BMDCs without or with VSV or IAV infection determined in immunoblots. Cell lysates (Lys); cell supernatants (Sup); error bars, SD; **, p<0.01; ***, p<0.001 by two-tailed Student’s t tests. See also Figure S2.
Figure 3. Involvement of NLRP3, ASC, and MAVS in IL-1β induction but not TNFα induction in response to viral infections
(A,B) LPS primed wt, Nlrp3−/− and Asc−/− BMDC were infected with VSV or IAV. (C,D) LPS primed wt, Rig I−/−, _Mda5_−/−, RigI_−/−/Mda5_−/−, and Mavs−/− BMDC were infected with VSV or IAV. IL-1β and TNFα levels were measured by ELISAs. Error bars, SD; ***, p<0.001 by two-tailed Student’s t tests. See also Figure S3.
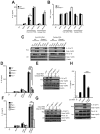
Figure 4. IL-1β production by direct activation of RNase L is dependent on both NLRP3 and ASC
BMDC were primed with Pam3Csk4 (200 ng/ml) for 16 hrs and mock transfected or transfected with (2′-5′)p3A3 for additional 8 hrs as indicated. (A) rRNA cleavage in response to (2′-5′)p3A3 transfection was monitored in RNA chips. (B,C) IL-1β and cleaved caspase-1 was measured in cell supernatants (Sup), whereas proIL-1β, procaspase-1 and β-actin were measured in cell lysates (Lys). Upper panels, ELISAs; lower panels, immunoblots. Error bars, SD; ***; p<0.001 by two-tailed Student’s t tests.
Figure 5. RNase L generated RNA cleavage products bearing 2′,3′-cyclic phosphates induce IL-1β production
(A–C) wt and Rnasel−/− BMDC were primed with Pam3Csk4 (200 ng/ml) for 16 hrs and transfected with intact or RNase L-cleaved IAV RNA or cellular RNA for 8 hrs. (D–G) BMDC of different genotypes (as indicated) were primed with Pam3Csk4 and transfected with uncleaved or RNase L-cleaved cellular RNAs for 8 hrs. (H) Intact, cleaved, or cleaved and PNK-treated cellular RNA were transfected into Pam3Csk4 treated BMDC. (A,B,D,F,&H upper) ELISAs; (C,E,G, and H lower) immunoblots. Cell supernatants (Sup); cell lysates (Lys). Error bars, SD;. **p<0.01;***, p<0.001 by two-tailed Student’s t tests. See also Figure S4.
Figure 6. A functional nuclease domain in RNase L is required for inflammasome stimulation
(A) RNase L levels in THP-1 cells or expressing sh-Rnasel alone or with Flag-tagged wt or R667A mutant RNase L cDNAs. Immunoblots were probed with (upper panel) monoclonal antibody to human RNase L, (middle panel) antibody to Flag epitope, and (bottom panel) antibody to β-actin. (B) IL-1β and caspase-1 (p20) in cell supernatants (Sup) and procaspase-1 (p45) and β-actin in cell lysates (Lys) from THP-1 cells expressing sh-control or sh-Rnasel and infected with IAV, IAV/ΔNS1 or VSV. (C,D) THP-1 cells expressing Rnasel shRNA or reconstituted with wt or mutant R667A RNase L infected with (C) IAV/ΔNS1 or (D) VSV. (B–D), upper panels, ELISAs; lower panels, immunoblots. Error bars, SD; ***, p<0.001 by two-tailed Student’s t tests. See also Figure S5.
Figure 7. Involvement of DHX33 in RNase L-mediated inflammasome activation
(A) shRNA mediated depletion of DDX1 or DHX33 in THP-1 macrophages. (B–E) THP-1 macrophages expressing different shRNAs [sh-control, sh-Ddx1 or sh-_Dhx33_] were (B) transfected with pIC (2 μg/ml for 8 hrs), (C) infected with VSV, (D) infected with IAV/ΔNS1, and (E) mock transfected or transfected with cell RNA cleaved with RNase L or transfected with (2′-5′)p3A3. Upper panels, ELISAs; lower panels, immunoblots (IB). (F) THP-1 macrophages were transfected with intact, cleaved, cleaved and PNK-treated cellular RNA or pIC. Immunoprecipitations (IP) were with anti-NLRP3, anti-DHX33, or an isotype control IgG. Cell lysates were used as controls (bottom panel). (G) HEK293T cells were co-transfected with myc-MAVS and flag-NLRP3 cDNAs. After 72 hrs, cells were transfected with intact, cleaved or cleaved and PNK treated cellular RNA. IPs were with anti-myc followed by immunoblotting (IB) with anti-flag, anti-DHX33 or anti-myc antibodies. Cell lysates were used as controls (lower panel). (H) Purified flag-DHX33 was incubated with biotinylated IAV M gene RNA (intact, cleaved, or cleaved and PNK-treated). Biotinylated RNAs were precipitated with streptavidin beads followed by IB with anti-flag antibody to detect DHX33. cell supernatants (Sup); cell lysates (lys). Error bars, SD; **p<0.01;***, p<0.001 by two-tailed Student’s t tests. See also Figure S6.
Similar articles
- RNase L and the NLRP3-inflammasome: An old merchant in a new trade.
Banerjee S. Banerjee S. Cytokine Growth Factor Rev. 2016 Jun;29:63-70. doi: 10.1016/j.cytogfr.2016.02.008. Epub 2016 Mar 10. Cytokine Growth Factor Rev. 2016. PMID: 26987611 - NS1 Protein of 2009 Pandemic Influenza A Virus Inhibits Porcine NLRP3 Inflammasome-Mediated Interleukin-1 Beta Production by Suppressing ASC Ubiquitination.
Park HS, Liu G, Thulasi Raman SN, Landreth SL, Liu Q, Zhou Y. Park HS, et al. J Virol. 2018 Mar 28;92(8):e00022-18. doi: 10.1128/JVI.00022-18. Print 2018 Apr 15. J Virol. 2018. PMID: 29386291 Free PMC article. - The RNA- and TRIM25-Binding Domains of Influenza Virus NS1 Protein Are Essential for Suppression of NLRP3 Inflammasome-Mediated Interleukin-1β Secretion.
Moriyama M, Chen IY, Kawaguchi A, Koshiba T, Nagata K, Takeyama H, Hasegawa H, Ichinohe T. Moriyama M, et al. J Virol. 2016 Mar 28;90(8):4105-4114. doi: 10.1128/JVI.00120-16. Print 2016 Apr. J Virol. 2016. PMID: 26865721 Free PMC article. - The role of the NLRP3 inflammasome in regulation of antiviral responses to influenza A virus infection.
Sarvestani ST, McAuley JL. Sarvestani ST, et al. Antiviral Res. 2017 Dec;148:32-42. doi: 10.1016/j.antiviral.2017.10.020. Epub 2017 Oct 31. Antiviral Res. 2017. PMID: 29097227 Review. - Regulation and Function of the Nucleotide Binding Domain Leucine-Rich Repeat-Containing Receptor, Pyrin Domain-Containing-3 Inflammasome in Lung Disease.
Lee S, Suh GY, Ryter SW, Choi AM. Lee S, et al. Am J Respir Cell Mol Biol. 2016 Feb;54(2):151-60. doi: 10.1165/rcmb.2015-0231TR. Am J Respir Cell Mol Biol. 2016. PMID: 26418144 Free PMC article. Review.
Cited by
- RNA helicase SKIV2L limits antiviral defense and autoinflammation elicited by the OAS-RNase L pathway.
Yang K, Dong B, Asthana A, Silverman RH, Yan N. Yang K, et al. EMBO J. 2024 Sep;43(18):3876-3894. doi: 10.1038/s44318-024-00187-1. Epub 2024 Aug 7. EMBO J. 2024. PMID: 39112803 Free PMC article. - Research progress on the nonstructural protein 1 (NS1) of influenza a virus.
Zhang X, Zhang Y, Wei F. Zhang X, et al. Virulence. 2024 Dec;15(1):2359470. doi: 10.1080/21505594.2024.2359470. Epub 2024 Jun 25. Virulence. 2024. PMID: 38918890 Free PMC article. Review. - Endonucleolytic RNA cleavage drives changes in gene expression during the innate immune response.
Karasik A, Lorenzi HA, DePass AV, Guydosh NR. Karasik A, et al. Cell Rep. 2024 Jun 25;43(6):114287. doi: 10.1016/j.celrep.2024.114287. Epub 2024 May 31. Cell Rep. 2024. PMID: 38823018 Free PMC article. - Interferon signaling in the nasal epithelium distinguishes among lethal and common cold coronaviruses and mediates viral clearance.
Otter CJ, Renner DM, Fausto A, Tan LH, Cohen NA, Weiss SR. Otter CJ, et al. Proc Natl Acad Sci U S A. 2024 May 21;121(21):e2402540121. doi: 10.1073/pnas.2402540121. Epub 2024 May 17. Proc Natl Acad Sci U S A. 2024. PMID: 38758698 - Initiation of a ZAKα-dependent ribotoxic stress response by the innate immunity endoribonuclease RNase L.
Xi J, Snieckute G, Martínez JF, Arendrup FSW, Asthana A, Gaughan C, Lund AH, Bekker-Jensen S, Silverman RH. Xi J, et al. Cell Rep. 2024 Apr 23;43(4):113998. doi: 10.1016/j.celrep.2024.113998. Epub 2024 Mar 28. Cell Rep. 2024. PMID: 38551960 Free PMC article.
References
- Carroll SS, Chen E, Viscount T, Geib J, Sardana MK, Gehman J, Kuo LC. Cleavage of oligoribonucleotides by the 2′,5′-oligoadenylate- dependent ribonuclease L. The Journal of biological chemistry. 1996;271:4988–4992. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA044059/CA/NCI NIH HHS/United States
- U19 AI083019/AI/NIAID NIH HHS/United States
- AI88778/AI/NIAID NIH HHS/United States
- R01 DK091191/DK/NIDDK NIH HHS/United States
- R01AI063331/AI/NIAID NIH HHS/United States
- R01 AI074973/AI/NIAID NIH HHS/United States
- AI083019/AI/NIAID NIH HHS/United States
- U19 AI088778/AI/NIAID NIH HHS/United States
- AI074973/AI/NIAID NIH HHS/United States
- R01 AI063331/AI/NIAID NIH HHS/United States
- R01DK091191/DK/NIDDK NIH HHS/United States
- R01 CA044059/CA/NCI NIH HHS/United States
- R01 AI104002/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous