Genomic prediction of traits related to canine hip dysplasia - PubMed (original) (raw)
Genomic prediction of traits related to canine hip dysplasia
Enrique Sánchez-Molano et al. Front Genet. 2015.
Abstract
Increased concern for the welfare of pedigree dogs has led to development of selection programs against inherited diseases. An example is canine hip dysplasia (CHD), which has a moderate heritability and a high prevalence in some large-sized breeds. To date, selection using phenotypes has led to only modest improvement, and alternative strategies such as genomic selection (GS) may prove more effective. The primary aims of this study were to compare the performance of pedigree- and genomic-based breeding against CHD in the UK Labrador retriever population and to evaluate the performance of different GS methods. A sample of 1179 Labrador Retrievers evaluated for CHD according to the UK scoring method (hip score, HS) was genotyped with the Illumina CanineHD BeadChip. Twelve functions of HS and its component traits were analyzed using different statistical methods (GBLUP, Bayes C and Single-Step methods), and results were compared with a pedigree-based approach (BLUP) using cross-validation. Genomic methods resulted in similar or higher accuracies than pedigree-based methods with training sets of 944 individuals for all but the untransformed HS, suggesting that GS is an effective strategy. GBLUP and Bayes C gave similar prediction accuracies for HS and related traits, indicating a polygenic architecture. This conclusion was also supported by the low accuracies obtained in additional GBLUP analyses performed using only the SNPs with highest test statistics, also indicating that marker-assisted selection (MAS) would not be as effective as GS. A Single-Step method that combines genomic and pedigree information also showed higher accuracy than GBLUP and Bayes C for the log-transformed HS, which is currently used for pedigree based evaluations in UK. In conclusion, GS is a promising alternative to pedigree-based selection against CHD, requiring more phenotypes with genomic data to improve further the accuracy of prediction.
Keywords: Labrador Retrievers; dogs; genomic selection; hip dysplasia.
Figures
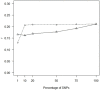
Figure 1
Impact of SNP density on prediction accuracy. GBLUP correlations (r) for THS for different numbers of SNP markers; markers were chosen at random (dashed line) or based on _p_-values from GWAS performed in the training populations (straight line). The number of analyzed SNPs (crosses) were 1% (1063), 10% (10,629), 20% (21,257), 50% (53,141), 75% (79,712), and 100% (106,282).
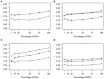
Figure 2
Comparison of correlations (r) for GWAS-markers. GBLUP was tested by estimating effects in the validation populations of the top SNPs identified by GWAS analysis performed in the training populations. Traits presented are (A) HS (circles) and THS (triangles). (B) NA_right (triangles), NA_left (circles), and NA_total (crosses). (C) SUB_right (triangles), SUB_left (circles), and SUB_total (crosses). (D) CrAE_right (triangles), CrAE_left (circles), and CrAE_total (crosses).
Similar articles
- Genomic Prediction of Two Complex Orthopedic Traits Across Multiple Pure and Mixed Breed Dogs.
Jiang L, Li Z, Hayward JJ, Hayashi K, Krotscheck U, Todhunter RJ, Tang Y, Huang M. Jiang L, et al. Front Genet. 2021 Sep 22;12:666740. doi: 10.3389/fgene.2021.666740. eCollection 2021. Front Genet. 2021. PMID: 34630503 Free PMC article. - Joint Genomic Prediction of Canine Hip Dysplasia in UK and US Labrador Retrievers.
Edwards SM, Woolliams JA, Hickey JM, Blott SC, Clements DN, Sánchez-Molano E, Todhunter RJ, Wiener P. Edwards SM, et al. Front Genet. 2018 Mar 28;9:101. doi: 10.3389/fgene.2018.00101. eCollection 2018. Front Genet. 2018. PMID: 29643866 Free PMC article. - Accuracy of genomic selection for a sib-evaluated trait using identity-by-state and identity-by-descent relationships.
Vela-Avitúa S, Meuwissen TH, Luan T, Ødegård J. Vela-Avitúa S, et al. Genet Sel Evol. 2015 Feb 25;47(1):9. doi: 10.1186/s12711-014-0084-2. Genet Sel Evol. 2015. PMID: 25888184 Free PMC article. - Selection against canine hip dysplasia: success or failure?
Wilson B, Nicholas FW, Thomson PC. Wilson B, et al. Vet J. 2011 Aug;189(2):160-8. doi: 10.1016/j.tvjl.2011.06.014. Epub 2011 Jul 2. Vet J. 2011. PMID: 21727013 Review. - Emerging insights into the genetic basis of canine hip dysplasia.
Ginja M, Gaspar AR, Ginja C. Ginja M, et al. Vet Med (Auckl). 2015 May 20;6:193-202. doi: 10.2147/VMRR.S63536. eCollection 2015. Vet Med (Auckl). 2015. PMID: 30101106 Free PMC article. Review.
Cited by
- Genomic analysis and prediction of genomic values for distichiasis in Staffordshire bull terriers.
Jørgensen D, Ropstad EO, Meuwissen T, Lingaas F. Jørgensen D, et al. Canine Med Genet. 2023 Jul 24;10(1):9. doi: 10.1186/s40575-023-00132-1. Canine Med Genet. 2023. PMID: 37488637 Free PMC article. - Genomic Prediction of Two Complex Orthopedic Traits Across Multiple Pure and Mixed Breed Dogs.
Jiang L, Li Z, Hayward JJ, Hayashi K, Krotscheck U, Todhunter RJ, Tang Y, Huang M. Jiang L, et al. Front Genet. 2021 Sep 22;12:666740. doi: 10.3389/fgene.2021.666740. eCollection 2021. Front Genet. 2021. PMID: 34630503 Free PMC article. - Methods to Improve Joint Genetic Evaluation of Canine Hip Dysplasia Across BVA/KC and FCI Screening Schemes.
Wang S, Friedrich J, Strandberg E, Arvelius P, Wiener P. Wang S, et al. Front Vet Sci. 2020 Aug 11;7:386. doi: 10.3389/fvets.2020.00386. eCollection 2020. Front Vet Sci. 2020. PMID: 32850996 Free PMC article. - Bayesian and Machine Learning Models for Genomic Prediction of Anterior Cruciate Ligament Rupture in the Canine Model.
Baker LA, Momen M, Chan K, Bollig N, Lopes FB, Rosa GJM, Todhunter RJ, Binversie EE, Sample SJ, Muir P. Baker LA, et al. G3 (Bethesda). 2020 Aug 5;10(8):2619-2628. doi: 10.1534/g3.120.401244. G3 (Bethesda). 2020. PMID: 32499222 Free PMC article. - Canine hip dysplasia screening: Comparison of early evaluation to final grading in 231 dogs with Fédération Cynologique Internationale A and B.
Merca R, Bockstahler B, Vezzoni A, Tichy A, Boano S, Vidoni B. Merca R, et al. PLoS One. 2020 May 18;15(5):e0233257. doi: 10.1371/journal.pone.0233257. eCollection 2020. PLoS One. 2020. PMID: 32421701 Free PMC article.
References
Grants and funding
- BB/H019073/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/R/00001618/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/K001744/1/MRC_/Medical Research Council/United Kingdom
- BBS/E/D/20211554/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/B/05710/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G0900740/MRC_/Medical Research Council/United Kingdom
- BBS/E/R/00001610/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/05191132/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/R/00000673/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/H017798/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/20211553/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources