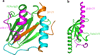Structural mechanism of integrin inactivation by filamin - PubMed (original) (raw)
Structural mechanism of integrin inactivation by filamin
Jianmin Liu et al. Nat Struct Mol Biol. 2015 May.
Abstract
Activation of heterodimeric (αβ) integrin is crucial for regulating cell adhesion. Binding of talin to the cytoplasmic face of integrin activates the receptor, but how integrin is maintained in a resting state to counterbalance its activation has remained obscure. Here, we report the structure of the cytoplasmic domain of human integrin αIIbβ3 bound to its inhibitor, the immunoglobin repeat 21 of filamin A (FLNa-Ig21). The structure reveals an unexpected ternary complex in which FLNa-Ig21 not only binds to the C terminus of the integrin β3 cytoplasmic tail (CT), as previously predicted, but also engages N-terminal helices of αIIb and β3 CTs to stabilize an inter-CT clasp that helps restrain the integrin in a resting state. Combined with functional data, the structure reveals a new mechanism of filamin-mediated retention of inactive integrin, suggesting a new framework for understanding regulation of integrin activation and adhesion.
Figures
Figure 1
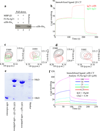
Interaction of FLNa-Ig21 with αIIbβ3 CTs. (a) Selected region of HSQC of 0.1mM 15N-labeled β3-CT in the absence (black) and presence (red) of 0.1mM FLNa-Ig21. Strongly perturbed residues are labeled in the spectrum of the free β3-CT. (b) Representative real time SPR sensorgrams of the binding between β3-CT and FLNa-Ig21 (n=2). The sensorgrams were fitted into a two-site binding model with KD1~4.9µM and KD2~150µM respectively. (c) Selected regions of HSQC of 0.1mM 15N-labeled αIIb-CT in the absence (black) and presence (red) of 0.1mM FLNa-Ig21. (d) Representative real-time SPR sensorgrams of the binding between αIIb-CT and FLNa-Ig21 (n=2). The real-time binding curves were fitted using a global fitting algorithm to a 1:1 binding model, resulting in the KD~229µM.
Figure 2
FLNa-Ig21 promotes the ternary complex formation with αIIb-CT and β3-CT. (a) Pull down experiment showing that while MBP-β3 failed to pull down His-tagged αIIb-CT, it did so in the presence of FLNa-Ig21; (b) Co-injection on β3 CT surface. 300µM FLNa-Ig21 followed by 300µM FLNa-Ig21 (green sensorgram). 300µM FLNa-Ig21 followed by 300µM FLNa-Ig21 plus 300µM αIIb CT (red sensorgram). n=2; (c) Selected regions of HSQC spectra of 0.1mM 15N-labeled αIIb-CT in the absence (black) and presence of 0.1mM FLNa-Ig21 (red), and presence of 0.1mM FLNa-Ig21 and 0.2mM β3-N; (d) Selected regions of HSQC spectra of 0.1mM 15N-labeled αIIb-CT in the absence (black) and presence of 0.1mM FLNa-Ig21-β3-CT chimera (red). (e) Pull down experiment showing that while GST-FLNa-Ig21 failed to pull down αIIb CT due to low affinity, GST-FLNa-Ig21-β3 CT chimera effectively pulled down αIIb CT. (f) SPR experiment showing that FLNa-Ig21-β3 CT binds to αIIb CT at KD~19µM. n=2.
Figure 3
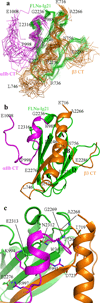
Structure of FLNa-Ig21-αIIb-CT-β3-CT ternary complex. (a) Superposition of 20 calculated FLNa-Ig21(green)-αIIb-CT(pink)-β3-CT(brown) complex structures with lowest energies, showing a well-defined structure. (b) Corresponding cartoon representation of the structure in (A) with the lowest energy. (c) Expanded regions of the complex interface based on (b).
Figure 4
A topology of FLNa-Ig21 bound to the αIIbβ3 CT heterodimer at the inner membrane surface. (a) Superposition of FLNa-Ig21-αIIbβ3-CT complex with αIIbβ3 transmembrane-cytoplasmic heterodimer (PDB code 2KNC) showing a cluster of positively charged residues H2239, K2240, and R2242 from FLNa-Ig21, αIIb-CT K989, and β3-CT K716 at the transmembrane-cytoplasmic border; (b). Selected regions of HSQC of 0.1mM 15N-labeled FLNa-Ig21 in the absence (black) and presence (red) of 1mM LUV vesicle showing the selective perturbation of the membrane binding site of FLNa-Ig21 involving the highlighted residues in Fig. 4A; (c) HSQC data indicating that FLNa-Ig21 K2240A mutation disrupts the FLNa-Ig21-membrane interaction. (d) Selective M987 signal from HSQC spectra of 0.1mM 15N-labeled αIIb TMCD bound to unlabeled β3 TMCD (1:1) in the absence (black) and presence (red) of equimolar FLNa-Ig21. The spectra were collected in 50mM POPC/POPS/DHPC (q=0.3) bicelle in 25mM HEPES, 5%(v/v) D2O, 0.02% NaN3, pH 7.4. (e). Selective G991 peak from the same HSQC spectra in (d) changed in the same trend as M987 in (d). (f). The intensity ratio (complex/free form) of M987 and G991 from the HSQC spectra of (d) and (e) in the absence (red) and presence of FLNa-Ig21.
Figure 5
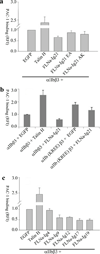
Functional evidence of FLNa-Ig21-mediated inhibition on integrin activation. Data are normalized for differences in surface expression of αIIbβ3 integrin as measured by 2G12 antibody. In all experiments, EGFP or EGFP-tagged proteins are transiently expressed with αIIbβ3. RFI values are Mean±S.E.M of fold change in PAC-1 binding over transiently expressed αIIbβ3 and EGFP (scaled to 1), from multiple independent experiments. Error bars in all experiments represent standard error of mean. (a) Integrin activation assay showing the different effects of talin head (talin-H), FLNa-Ig21, and the FLNa-Ig21 mutants (E2276A, F21EA; A2268K, F21AK) on the activation state of the transiently expressed αIIbβ3 (n=4). For each experiment, 3 transfections were set up that were processed independently as 3 biological replicants. The difference between vector (EGFP) and FLNa-Ig21 is significant (p<0.001). The difference between vector (EGFP) and E2276A (F21EA) or A2268K (F21AK) is not significant (p>0.05). (b) Integrin activation assay showing that transiently expressed αIIb-K994E-R997E-β3 mutant, αIIb(KREE)β3, exhibits substantially enhanced PAC-1 binding as compared to the WT integrin (p<0.05). N=2; 2 transfections for each condition were set up that were processed independently as 3 biological replicates. (c). Integrin activation assay showing that FLNa-Ig9 (p<0.05), Ig12 (p<0.001), Ig17 (p<0.001), Ig19 (p<0.001) exert inhibitory effect on the PAC-1 binding of the transiently expressed αIIbβ3. FLNa-Ig4 did not have significant inhibitory effect (p>0.05). n=2; 2 transfections for each condition were set up that were processed independently as 3 biological replicates.
Figure 6
Comparison of FLNa-Ig21 bound integrin αIIbβ3 CT complex (a) with talin-F3 bound β1 CT (b) (derived from PDB code 3G9W) showing how FLNa-Ig21 may compete with both β3-MP and β3-C for binding to talin-F3. Note that the C-terminus of β CT is β-strand (brown) when bound to FLNa-Ig21 (a) versus an extended loop when bound to talin-F3 (b), and thus the two binding events are mutually exclusive. Also binding of talin-F3 with β3-MP (yellow helix) in (b) would also cause steric clash with FLNa-Ig21 bound to the β3-MP (Cyan + Brown) in (a). The helical region in brown is involved in the interaction with FLNa-Ig21.
Comment in
- Integrin bondage: filamin takes control.
De Franceschi N, Ivaska J. De Franceschi N, et al. Nat Struct Mol Biol. 2015 May;22(5):355-7. doi: 10.1038/nsmb.3024. Nat Struct Mol Biol. 2015. PMID: 25945885 No abstract available.
Similar articles
- A mechanism of platelet integrin αIIbβ3 outside-in signaling through a novel integrin αIIb subunit-filamin-actin linkage.
Liu J, Lu F, Ithychanda SS, Apostol M, Das M, Deshpande G, Plow EF, Qin J. Liu J, et al. Blood. 2023 May 25;141(21):2629-2641. doi: 10.1182/blood.2022018333. Blood. 2023. PMID: 36867840 Free PMC article. - Binary and ternary complexes of FLNa-Ig21 with cytosolic tails of αMß2 integrin reveal dual role of filamin mediated regulation.
Zhiping LL, Ong LT, Chatterjee D, Tan SM, Bhattacharjya S. Zhiping LL, et al. Biochim Biophys Acta Gen Subj. 2021 Dec;1865(12):130005. doi: 10.1016/j.bbagen.2021.130005. Epub 2021 Sep 9. Biochim Biophys Acta Gen Subj. 2021. PMID: 34509570 - NMR analysis of the alphaIIb beta3 cytoplasmic interaction suggests a mechanism for integrin regulation.
Metcalf DG, Moore DT, Wu Y, Kielec JM, Molnar K, Valentine KG, Wand AJ, Bennett JS, DeGrado WF. Metcalf DG, et al. Proc Natl Acad Sci U S A. 2010 Dec 28;107(52):22481-6. doi: 10.1073/pnas.1015545107. Epub 2010 Dec 14. Proc Natl Acad Sci U S A. 2010. PMID: 21156831 Free PMC article. - Cytoplasmic binding partners of the platelet integrin alphaIIb beta3.
Bonner MA, Naik UP. Bonner MA, et al. Front Biosci. 2007 Jan 1;12:2038-49. doi: 10.2741/2209. Front Biosci. 2007. PMID: 17127442 Review. - Talin-dependent integrin signalling in vivo.
Petrich BG. Petrich BG. Thromb Haemost. 2009 Jun;101(6):1020-4. Thromb Haemost. 2009. PMID: 19492142 Review.
Cited by
- Charting the importance of filamin A posttranslational modifications.
Shead KD, Salyahetdinova V, Baillie GS. Shead KD, et al. Biochem J. 2024 Jul 3;481(13):865-881. doi: 10.1042/BCJ20240121. Biochem J. 2024. PMID: 38958472 Free PMC article. Review. - A novel replication initiation region encoded in a widespread Acinetobacter plasmid lineage carrying a blaNDM-1 gene.
Bello-López E, Pérez-Oseguera Á, Santos W, Cevallos MÁ. Bello-López E, et al. PLoS One. 2024 May 31;19(5):e0303976. doi: 10.1371/journal.pone.0303976. eCollection 2024. PLoS One. 2024. PMID: 38820537 Free PMC article. - PACSIN2 regulates platelet integrin β1 hemostatic function.
Biswas R, Boyd EK, Eaton N, Steenackers A, Schulte ML, Reusswig F, Yu H, Drew C, Kahr WHA, Shi Q, Plomann M, Hoffmeister KM, Falet H. Biswas R, et al. J Thromb Haemost. 2023 Dec;21(12):3619-3632. doi: 10.1016/j.jtha.2023.08.026. Epub 2023 Sep 9. J Thromb Haemost. 2023. PMID: 37678551 Free PMC article. - A novel association between platelet filamin A and soluble N-ethylmaleimide sensitive factor attachment proteins regulates granule secretion.
Golla K, Paul M, Lengyell TC, Simpson EM, Falet H, Kim H. Golla K, et al. Res Pract Thromb Haemost. 2022 Dec 20;7(4):100019. doi: 10.1016/j.rpth.2022.100019. eCollection 2023 May. Res Pract Thromb Haemost. 2022. PMID: 37538498 Free PMC article. - Cryo-EM structures of full-length integrin αIIbβ3 in native lipids.
Adair BD, Xiong JP, Yeager M, Arnaout MA. Adair BD, et al. Nat Commun. 2023 Jul 13;14(1):4168. doi: 10.1038/s41467-023-39763-0. Nat Commun. 2023. PMID: 37443315 Free PMC article.
References
- Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110(6):673–687. - PubMed
- Bouvard D, Pouwels J, De Franceschi N, Ivaska J. Integrin inactivators: balancing cellular functions in vitro and in vivo. Nat Rev Mol Cell Biol. 2013;14(7):430–442. - PubMed
- Kiema T, et al. The molecular basis of filamin binding to integrins and competition with talin. Mol Cell. 2006;21(3):337–347. - PubMed
- Stossel TP, et al. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001;2(2):138–145. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R56 DK102020/DK/NIDDK NIH HHS/United States
- P01 HL073311/HL/NHLBI NIH HHS/United States
- DK102020/DK/NIDDK NIH HHS/United States
- R01 GM062823/GM/NIGMS NIH HHS/United States
- R01 DK102020/DK/NIDDK NIH HHS/United States
- UL1 TR000439/TR/NCATS NIH HHS/United States
- R01 HL096062/HL/NHLBI NIH HHS/United States
- HL073311/HL/NHLBI NIH HHS/United States
- GM062823/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous