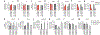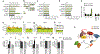A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature - PubMed (original) (raw)
A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature
Inma Gonzalez et al. Nat Struct Mol Biol. 2015 May.
Abstract
Alternative pre-mRNA splicing is a highly cell type-specific process essential to generating protein diversity. However, the mechanisms responsible for the establishment and maintenance of heritable cell-specific alternative-splicing programs are poorly understood. Recent observations point to a role of histone modifications in the regulation of alternative splicing. Here we report a new mechanism of chromatin-mediated splicing control involving a long noncoding RNA (lncRNA). We have identified an evolutionarily conserved nuclear antisense lncRNA, generated from within the human FGFR2 locus, that promotes epithelial-specific alternative splicing of FGFR2. The lncRNA acts through recruitment of Polycomb-group proteins and the histone demethylase KDM2a to create a chromatin environment that impairs binding of a repressive chromatin-splicing adaptor complex important for mesenchymal-specific splicing. Our results uncover a new function for lncRNAs in the establishment and maintenance of cell-specific alternative splicing via modulation of chromatin signatures.
Figures
Figure 1
Polycomb-complex components in FGFR2 alternative-splicing regulation. (a—l) Recruitment of EZH2 (a,b), SUZ12 (c,d), CBX8 (e,f), HP1γ (g,h), KDM2a (i,j) and KDM2b (k,l) to _FGFR2_and control CD44 loci in PNT2 and hMSC cells. ChIP values (mean ± s.e.m.; n = 3 independent experiments) are depicted as percentage of input, with no-antibody (no Ab) ChIP as negative control. *P < 0.05; **P < 0.01 by two-tailed Student’s t test relative to hMSCs. TSS, transcription start site. (m-v) Inclusion levels of FGFR2 exons Illb (m,r) and Illc (n,s), and control CD44-v6 (p,u) and HMGA1-e2 (q,v), in PNT2 and hMSC cells after expression of EZH2 and PHF1 (m-q) or KDM2a (r-v). qRT-PCR values (mean ± s.e.m.; n = 9 independent experiments) are normalized to total mRNA of the corresponding gene and are shown as the fold change over transfection with empty vector (mock). **P < 0.001 by two-tailed Student’s t test relative to mock transfection. IIIb/IIIc, splicing ratio of exons IIIb and IIIc (o,t).
Figure 2
EZH2 promotes _FGFR2_exon Illb inclusion via KDM2a. (a-l) Levels of EZH2 (a,b), H3K27me3 (c,d), KDM2a (e,f), H3K36me2 (g,h,k,l) and MRG15 (i,j) along FGFR2 and control CD44 loci after combined expression of EZH2 and PHF1 (a-h) or KDM2a (i-l) in hMSCs. ChIP values (mean ± s.e.m.; n = 4 independent experiments) are shown as the percentage of input or normalized to control H3. * P < 0.05; **P < 0.01 by two-tailed Student’s t test relative to mock transfections. (m-s) Inclusion levels of FGFR2 exons Illb (m) and lllc (n), and controls CD44-v6 (p), HMGA1-e2 (q), KDM2A (r) and EZH2 (s) after expression of EZH2 in the presence or absence of KDM2a in hMSCs. The gene encoding cyclophilin A (CycA; official symbol PPIA) is a housekeeping control. qRT-PCR values (mean ± s.e.m.; n = 6 independent experiments) are normalized to total mRNA of the corresponding gene and are shown as fold change over transfection with negative siRNA and empty vector (mock). **P < 0.01 by two-tailed Student’s t test relative to mock. Illb/lllc, splicing ratio of exons Illb and lllc (o).
Figure 3
A IncRNA antisense to FGFR2 promotes exon Illb inclusion. (a) UCSC Genome Browser mapping of the FGFR2 locus (blue) and the full sequence of asFGFR2 lncRNA identified by 5’−3’ RACE (orange). Red, northern blot probes that detected as FGFR2; black, negative probes. Chr, chromosome. (b,c) Northern blot detection of asFGFR2 by exon lllc antisense probe (as(lllc)) in cells that include exon lllb (PNT2 and MCF7) and cells that do not include exon lllb (CRL, PRC30 and hMSCs). Detection of GAPDH and an FGFR2 sense intronic sequence (sense(i8)) are used as controls. From a nonradiolabeled RNA ladder (M, marker), the asFGFR2 fragment is estimated to be around 1 kb, in agreement with results obtained by RACE (Supplementary Data Set 1). (d-i) Expression levels of FGFR2 exons lllb (d) and lllc (e), and constitutive e14 (g), control CD44-v6 (h) and asFGFR2 after knockdown of asFGFR2 (anti-lllc) with antisense LNA-modified DNA oligonucleotides in PNT2 cells for 72 h. qRT-PCR values (mean ± s.e.m.; n = 6 independent experiments) are normalized to total mRNA and are shown as fold change over transfection with control siRNA. ln the case of asFGFR2, strand-specific qRT-PCR values (ssRT (i), mean ± s.e.m.; n = 4 independent experiments) are shown as the fold change over reverse transcription with no primer. Oligo, oligonucleotide. *P < 0.05; **P < 0.01 by two-tailed Student’s t test relative to control. (j-n) lnclusion levels of FGFR2 exons lllb (j), lllc (k), e14 (m) and CD44-v6 (n) after expression of asFGFR2 or an antisense GAPDH sequence in hMSC cells. qRT-PCR values (mean ± s.e.m.; n = 8 independent experiments) are normalized to total mRNA and are shown as the fold change over transfection with empty vector (mock). **P < 0.001 by two-tailed Student’s t test relative to mock. lllb/lllc, splicing ratio of exons lllb and lllc (f,l).
Figure 4
asFGFR2 effect on FGFR2 splicing is independent of Argonaute and Dicer. (a-g) Inclusion levels of FGFR2 exons Illb (a) and IIIc (b), after combined downregulation of Argonautes 1 and 2 (Ago siRNA) for 72 h in PNT2 (red) and hMSC (black) cells. The inclusion levels of the Ago-independent PKM2 (d) and the Ago-dependent PCSK6 (e) alternatively spliced exons are shown as controls. Knockdown validation of AGO1 (f) and AGO2 (g) are also shown. qRT-PCR values (mean ± s.e.m.; n = 6 independent experiments) are normalized to total mRNA of the corresponding gene and are shown as the fold change over transfection with a negative-control siRNA (Ctrl). * P < 0.01; **P < 0.001 by two-tailed Student’s t test relative to control siRNA. IIIb/IIIc, splicing ratio of exons IIIb and IIIc (c). (h-m) Splicing ratio of FGFR2 exons IIIb and IIIc (h), and expression levels of control CD44-v6 (i), asFGFR2 (j), DICER1 (k), AGO1 (l) and AGO2 (m) after combined overexpression of asFGFR2 and downregulation of DICER1 or AGO1 and AGO2 (Ago siRNAs) for 72 h in hMSC cells. qRT-PCR values (mean ± s.e.m.; n = 6 independent experiments) are normalized to cyclophilin A (CycA) or total mRNA and are shown as fold change over transfection with a control siRNA and asGAPDH. **P < 0.001 by two-tailed Student’s t test relative to cells treated with control siRNA and asGAPDH.
Figure 5
asFGFR2 induces an epithelial-specific chromatin signature in FGFR2. (a-n) Levels of SUZ12 (a,b,e,f), H3K27me3 (c,d,g,h), KDM2a (i j), H3K36me2 (k,l) and MRG15 (m,n) along FGFR2 and control CD44 loci after downregulation of asFGFR2 in PNT2 cells (a-d) or ectopic expression of asFGFR2 in hMSCs (e-n). ChIP values (mean ± s.e.m.; n = 3 independent experiments for downregulation and n = 4 for ectopic expression) are shown as the percentage of input or normalized to control H3. *P < 0.05; **P < 0.01 by two-tailed Student’s t test relative to asGAPDH or control siRNA. (o,p) RNA immunoprecipitation of PTB to exon IIIb pre-mRNA upon asFGFR2 expression in hMSCs (o) or SUZ12 to endogenous asFGFR2 in PNT2 cells (p). Exon e15 and cyclophilin A (CycA) are negative controls. qRT-PCR values (mean ± s.e.m.; n = 3 independent experiments) are the percentage of input relative to IgG RNA immunoprecipitation. *P < 0.05 by two-tailed Student’s t test relative to overexpression of asGAPDH.
Figure 6
asFGFR2’s effect on alternative splicing is dependent on the IIIc region and KDM2a. (a-d) Inclusion levels of FGFR2 exons Illb (a) and lllc (c), and control CD44-v6 (d) after expression of deletion mutants of asFGFR2 (b) for 48 h in hMSCs. qRT-PCR values (mean ± s.e.m.; n = 9 independent experiments) are normalized to their corresponding gene and are shown as the fold change over transfection with control asGAPDH. *P< 0.05; **P < 0.001 by two-tailed Student’s _t_test relative to asGAPDH. (e-h) Enrichment levels of SUZ12 (e,f) and H3K27me3 (g,h) along FGFR2 (e,g) and control CD44 (f,h) loci after expression of asFGFR2 mutants that lack antisense exons lllc (del(l I Ic+111 b) or that express only antisense lllc (as(111c+ Illb). Expression of asGAPDH is a control. ChIP values (mean ± s.e.m.; n = 3 independent experiments) are normalized to control H3. *P < 0.05; **P < 0.01 by two-tailed Student’s _t_test relative to del(lllc+lllb). (i-m) Inclusion levels of FGFR2 exons Illb (i) and lllc (j), e14 (l) and control CD44-v6 (m) after expression of asFGFR2 in the presence or absence of KDM2a in hMSC cells. qRT-PCR values (mean ± s.e.m.; n = 6, independent experiments) are normalized to total mRNA and are shown as the fold change of transfection with control siRNA and asGAPDH. *P < 0.05; **P < 0.01 by two-tailed Student’s _t_test relative to asGAPDH. Illb/lllc, splicing ratio of exons Illb and lllc (k). (n) Enrichment levels of the in vitro-transcribed biotinylated asFGFR2 along the FGFR2 locus in hMSC cells transfected with the biotinylated lncRNAs. Pulldown of a biotinylated asGAPDH sequence is a control. lncRNA pulldown values (mean ± s.e.m.; n = 4 independent experiments) are depicted as the percentage of input. (o) Schematic model of how expression of asFGFR2 (thick red line) induces recruitment in cis of PRC2 (EZH2 and SUZ12) to lead to enrichment of the H3K36 demethylase KDM2a, which inhibits recruitment of the chromatin-adaptor complex MRG15-PTB, thus favoring exon Illb inclusion.
Similar articles
- Argonaute proteins couple chromatin silencing to alternative splicing.
Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau JC, Mathieu J, Hamiche A, Ait-Si-Ali S, Muchardt C, Batsché E, Harel-Bellan A. Ameyar-Zazoua M, et al. Nat Struct Mol Biol. 2012 Oct;19(10):998-1004. doi: 10.1038/nsmb.2373. Epub 2012 Sep 9. Nat Struct Mol Biol. 2012. PMID: 22961379 - Regulation of alternative splicing by histone modifications.
Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Luco RF, et al. Science. 2010 Feb 19;327(5968):996-1000. doi: 10.1126/science.1184208. Epub 2010 Feb 4. Science. 2010. PMID: 20133523 Free PMC article. - Chromatin: the final frontier in splicing regulation?
Fox-Walsh K, Fu XD. Fox-Walsh K, et al. Dev Cell. 2010 Mar 16;18(3):336-8. doi: 10.1016/j.devcel.2010.03.002. Dev Cell. 2010. PMID: 20230741 Free PMC article. - Epigenetic regulation in human melanoma: past and future.
Sarkar D, Leung EY, Baguley BC, Finlay GJ, Askarian-Amiri ME. Sarkar D, et al. Epigenetics. 2015;10(2):103-21. doi: 10.1080/15592294.2014.1003746. Epigenetics. 2015. PMID: 25587943 Free PMC article. Review. - [The epigenetic effect on pre-mRNA alternative splicing].
Zhao J, Wang F, Xu Z, Fan Y. Zhao J, et al. Yi Chuan. 2014 Mar;36(3):248-55. Yi Chuan. 2014. PMID: 24846965 Review. Chinese.
Cited by
- LncSTPred: a predictive model of lncRNA subcellular localization and decipherment of the biological determinants influencing localization.
Hu SL, Chen YL, Zhang LQ, Bai H, Yang JH, Li QZ. Hu SL, et al. Front Mol Biosci. 2024 Sep 5;11:1452142. doi: 10.3389/fmolb.2024.1452142. eCollection 2024. Front Mol Biosci. 2024. PMID: 39301172 Free PMC article. - HULC: an oncogenic long non-coding RNA in human cancer.
Yu X, Zheng H, Chan MT, Wu WK. Yu X, et al. J Cell Mol Med. 2017 Feb;21(2):410-417. doi: 10.1111/jcmm.12956. Epub 2016 Oct 25. J Cell Mol Med. 2017. PMID: 27781386 Free PMC article. Review. - BRR2a Affects Flowering Time via FLC Splicing.
Mahrez W, Shin J, Muñoz-Viana R, Figueiredo DD, Trejo-Arellano MS, Exner V, Siretskiy A, Gruissem W, Köhler C, Hennig L. Mahrez W, et al. PLoS Genet. 2016 Apr 21;12(4):e1005924. doi: 10.1371/journal.pgen.1005924. eCollection 2016 Apr. PLoS Genet. 2016. PMID: 27100965 Free PMC article. - Functional roles of conserved lncRNAs and circRNAs in eukaryotes.
Li J, Wang X. Li J, et al. Noncoding RNA Res. 2024 Jun 25;9(4):1271-1279. doi: 10.1016/j.ncrna.2024.06.014. eCollection 2024 Dec. Noncoding RNA Res. 2024. PMID: 39036601 Free PMC article. Review. - Alternative splicing of neuronal genes: new mechanisms and new therapies.
Lipscombe D, Lopez Soto EJ. Lipscombe D, et al. Curr Opin Neurobiol. 2019 Aug;57:26-31. doi: 10.1016/j.conb.2018.12.013. Epub 2019 Jan 28. Curr Opin Neurobiol. 2019. PMID: 30703685 Free PMC article. Review.
References
- Pan Q, Shai O, Lee LJ, Frey BJ & Blencowe BJ Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40, 1413–1415 (2008). - PubMed
- Barash Y et al. Deciphering the splicing code. Nature 465, 53–59 (2010). - PubMed
- Alló M et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat. Struct. Mol. Biol. 16, 717–724 (2009). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous