Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor-derived extracellular vesicles - PubMed (original) (raw)
. 2015 May 10;6(13):11327-41.
doi: 10.18632/oncotarget.3598.
Sungyong You 1, Cristiana Spinelli 1, Samantha Morley 2 3, Mandana Zandian 1, Paul-Joseph Aspuria 4, Lorenzo Cavallini 1 5, Chiara Ciardiello 1 6, Mariana Reis Sobreiro 1, Matteo Morello 1, Geetanjali Kharmate 7, Su Chul Jang 8, Dae-Kyum Kim 8, Elham Hosseini-Beheshti 7, Emma Tomlinson Guns 7, Martin Gleave 7, Yong Song Gho 8, Suresh Mathivanan 9, Wei Yang 1, Michael R Freeman 1 2 3, Dolores Di Vizio 1 2 3
Affiliations
- PMID: 25857301
- PMCID: PMC4484459
- DOI: 10.18632/oncotarget.3598
Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor-derived extracellular vesicles
Valentina R Minciacchi et al. Oncotarget. 2015.
Abstract
Large oncosomes (LO) are atypically large (1-10 µm diameter) cancer-derived extracellular vesicles (EVs), originating from the shedding of membrane blebs and associated with advanced disease. We report that 25% of the proteins, identified by a quantitative proteomics analysis, are differentially represented in large and nano-sized EVs from prostate cancer cells. Proteins enriched in large EVs included enzymes involved in glucose, glutamine and amino acid metabolism, all metabolic processes relevant to cancer. Glutamine metabolism was altered in cancer cells exposed to large EVs, an effect that was not observed upon treatment with exosomes. Large EVs exhibited discrete buoyant densities in iodixanol (OptiPrep(TM)) gradients. Fluorescent microscopy of large EVs revealed an appearance consistent with LO morphology, indicating that these structures can be categorized as LO. Among the proteins enriched in LO, cytokeratin 18 (CK18) was one of the most abundant (within the top 5th percentile) and was used to develop an assay to detect LO in the circulation and tissues of mice and patients with prostate cancer. These observations indicate that LO represent a discrete EV type that may play a distinct role in tumor progression and that may be a source of cancer-specific markers.
Keywords: SILAC Proteomics; amoeboid blebbing; cancer metabolism; extracellular vesicles; tumor progression.
Conflict of interest statement
CONFLICTS OF INTEREST
D.D.V.: Inventor of two patents on EVs as circulating biomarkers of cancer (pending). M.R.F., V.R.M., M.M.: co-inventors of two patents on EVs as circulating biomarkers of cancer (pending).
Figures
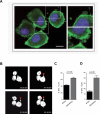
Figure 1. Silencing of DIAPH3 results in increased shedding of large EVs
(A) Membrane blebbing and shedding of EVs of variable size (insets) from DU145 cells stably expressing DIAPH3 shRNA, stained with CTxB-FITC (X 63). Scale bar, 20 μm. (B) 30 min-1h interval frames (from Supplementary Movie 1), acquired by real-time confocal microscopy of DIAPH3-silenced DU145 over-expressing RFP-tubulin. The arrow points to a membrane bleb that is released as a large oncosome. (C, D) EVs from DIAPH3-silenced or parental DU145 cells were analyzed by flow cytometry.
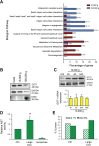
Figure 2. Identification of unique and differentially expressed proteins in large and nano-sized EVs
(A) Volcano plots of the log2 ratio of the averaged, normalized SILAC intensities against the FDR of the differential expression between large and nano-sized EVs. Red and blue dots correspond to proteins enriched in large EVs and nano-sized EVs, respectively. (B) Bar plots show the abundance of unique proteins quantified in large (right) and nano-sized EVs (left). The horizontal axis represents the normalized average log2 ion intensities of uniquely identified proteins.
Figure 3. Proteins highly abundant in large EVs are associated with cancer progression
Proteins with >4 fold enrichment in large EVs were functionally annotated by using the GO, KEGG and iHOP literature mining softwares to identify the association with cancer progression. The column on the right indicates the number of studies, obtained from the EVpedia database, in which these proteins were detected.
Figure 4. Large EVs are enriched in vesicular markers and alter glutamine metabolism in recipient cells
(A) Functional analysis using FunRich software indicates the biological pathways overrepresented either in large (10,000 g) or nano-sized EVs (100,000 g). (B) Protein lysates from cells, large and nano-sized EVs were blotted with the indicated antibodies. CD63 was expressed specifically in nano-sized EVs, and GOT1 in large EVs. (C) Protein lysates from DU145 cells untreated or treated with large EVs for the indicated times, were blotted with GOT1 antibody. GAPDH was used as a loading control (top panel). GOT1 mRNA expression levels in DU145 cells untreated or treated with large EVs for the indicated times do not exhibit significant changes. The result is displayed as levels of GOT1 transcript after normalization to the housekeeping gene GAPDH in treated versus untreated cells at 2-48h (bottom panel). (D) GOT1 activity was measured in DU145 cells in 5% glutamine with or without treatment with large oncosomes (1.15 g/ml OptiPrepTM density fractions) or exosomes (1.10 g/ml) (20μg/ml of protein lysate). The results from 3 experiments are displayed as relative AST activity in treated cells in comparison with the baseline activity of the enzyme (p=0.024). (E) Cell-cycle was analyzed by flow cytometry in DU145 cells treated with large oncosomes or vehicle in the presence of 1% or 5% glutamine for 24 hours.
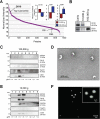
Figure 5. SILAC validation by OptiPrepTM gradient, EM and IF
(A) Rank plot of normalized ion intensities of all proteins identified in large EVs. CK18 is indicated as high abundant and CD9 and CD81 are indicated as low abundant proteins in large EVs. (B) Protein lysates from cells, large EVs and nano-sized EVs were blotted with the indicated antibodies. CD81 was expressed specifically in nano-sized EVs, and CK18 in large EVs. (C) Equal amounts of proteins (10 μg) from OptiPrepTM fractions (1-8) of nano-sized EVs were blotted with the indicated antibodies. Exosome markers CD81 and Tsg101 were identified in fraction 3, corresponding to the buoyant density of 1.10 g/ml. (D) Magnified TEM detail of negative stained EVs corresponding to the buoyant density of 1.10 g/ml, showing cup-shaped vesicles. Scale bars, 500nm. (E) Equal amounts of proteins (10 μg) from OptiPrepTM fractions (1-8) of large EVs were blotted with the indicated antibodies. Large EV enriched proteins (SILAC) such as CK18, GAPDH and HSPA5 were identified in fractions 3 and 4, corresponding to the buoyant density of 1.10-1.15 g/ml. (F) The 1.15 g/ml density fraction, labeled with DiO lipophilic dye, was imaged by IF. Scale bars, 10 μm and 2 μm (inset).
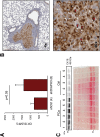
Figure 6. CK18 is a marker of large oncosomes in vivo
(A) FACS analysis of CK18 positive EVs from the plasma of mice with lung metastasis injected with DIAPH3-silenced (n=6) or control (n=4) DU145 cells. The plot shows the mean fluorescent intensity (MFI) relative to CK18 positive EVs >1 μm. (B) Representative lung tissue section immunostained with CK18 (4X). The tumor is strongly positive for CK18, and LO features can be identified at higher magnification (40X). (C) CK18 western blotting of EVs isolated by ExoQuickTM from the plasma of 6 patients with prostate cancer (PCa), and 5 healthy male subjects (Ctrl). Ponceau staining is displayed as a loading control.
Similar articles
- Large oncosomes overexpressing integrin alpha-V promote prostate cancer adhesion and invasion via AKT activation.
Ciardiello C, Leone A, Lanuti P, Roca MS, Moccia T, Minciacchi VR, Minopoli M, Gigantino V, De Cecio R, Rippa M, Petti L, Capone F, Vitagliano C, Milone MR, Pucci B, Lombardi R, Iannelli F, Di Gennaro E, Bruzzese F, Marchisio M, Carriero MV, Di Vizio D, Budillon A. Ciardiello C, et al. J Exp Clin Cancer Res. 2019 Jul 18;38(1):317. doi: 10.1186/s13046-019-1317-6. J Exp Clin Cancer Res. 2019. PMID: 31319863 Free PMC article. - Large oncosomes mediate intercellular transfer of functional microRNA.
Morello M, Minciacchi VR, de Candia P, Yang J, Posadas E, Kim H, Griffiths D, Bhowmick N, Chung LW, Gandellini P, Freeman MR, Demichelis F, Di Vizio D. Morello M, et al. Cell Cycle. 2013 Nov 15;12(22):3526-36. doi: 10.4161/cc.26539. Epub 2013 Sep 23. Cell Cycle. 2013. PMID: 24091630 Free PMC article. - Microvesicle- and exosome-mediated drug delivery enhances the cytotoxicity of Paclitaxel in autologous prostate cancer cells.
Saari H, Lázaro-Ibáñez E, Viitala T, Vuorimaa-Laukkanen E, Siljander P, Yliperttula M. Saari H, et al. J Control Release. 2015 Dec 28;220(Pt B):727-37. doi: 10.1016/j.jconrel.2015.09.031. Epub 2015 Sep 24. J Control Release. 2015. PMID: 26390807 - The Emerging Role of Extracellular Vesicle-Mediated Drug Resistance in Cancers: Implications in Advanced Prostate Cancer.
Soekmadji C, Nelson CC. Soekmadji C, et al. Biomed Res Int. 2015;2015:454837. doi: 10.1155/2015/454837. Epub 2015 Oct 26. Biomed Res Int. 2015. PMID: 26587537 Free PMC article. Review. - Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes.
Minciacchi VR, Freeman MR, Di Vizio D. Minciacchi VR, et al. Semin Cell Dev Biol. 2015 Apr;40:41-51. doi: 10.1016/j.semcdb.2015.02.010. Epub 2015 Feb 23. Semin Cell Dev Biol. 2015. PMID: 25721812 Free PMC article. Review.
Cited by
- Extracellular Vesicles as Biomarkers in Cancer Immunotherapy.
Mathew M, Zade M, Mezghani N, Patel R, Wang Y, Momen-Heravi F. Mathew M, et al. Cancers (Basel). 2020 Sep 30;12(10):2825. doi: 10.3390/cancers12102825. Cancers (Basel). 2020. PMID: 33007968 Free PMC article. Review. - Intercellular Vesicular Transfer by Exosomes, Microparticles and Oncosomes - Implications for Cancer Biology and Treatments.
Jaiswal R, Sedger LM. Jaiswal R, et al. Front Oncol. 2019 Mar 6;9:125. doi: 10.3389/fonc.2019.00125. eCollection 2019. Front Oncol. 2019. PMID: 30895170 Free PMC article. Review. - Extracellular vesicles and chronic inflammation during HIV infection.
Pérez PS, Romaniuk MA, Duette GA, Zhao Z, Huang Y, Martin-Jaular L, Witwer KW, Théry C, Ostrowski M. Pérez PS, et al. J Extracell Vesicles. 2019 Nov 6;8(1):1687275. doi: 10.1080/20013078.2019.1687275. eCollection 2019. J Extracell Vesicles. 2019. PMID: 31998449 Free PMC article. Review. - Extracellular Vesicles under Oxidative Stress Conditions: Biological Properties and Physiological Roles.
Chiaradia E, Tancini B, Emiliani C, Delo F, Pellegrino RM, Tognoloni A, Urbanelli L, Buratta S. Chiaradia E, et al. Cells. 2021 Jul 12;10(7):1763. doi: 10.3390/cells10071763. Cells. 2021. PMID: 34359933 Free PMC article. Review. - Extracellular Vesicle-Based Detection of Pancreatic Cancer.
Verel-Yilmaz Y, Fernández JP, Schäfer A, Nevermann S, Cook L, Gercke N, Helmprobst F, Jaworek C, Pogge von Strandmann E, Pagenstecher A, Bartsch DK, Bartsch JW, Slater EP. Verel-Yilmaz Y, et al. Front Cell Dev Biol. 2021 Jul 23;9:697939. doi: 10.3389/fcell.2021.697939. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34368146 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- R00 CA131472/CA/NCI NIH HHS/United States
- R01 CA143777/CA/NCI NIH HHS/United States
- P01 CA098912/CA/NCI NIH HHS/United States
- R01 DK087806/DK/NIDDK NIH HHS/United States
- P50 CA092131/CA/NCI NIH HHS/United States
- R37 DK47556/DK/NIDDK NIH HHS/United States
- R01 CA112303/CA/NCI NIH HHS/United States
- NCI NIH R00 CA131472/CA/NCI NIH HHS/United States
- CA112303/CA/NCI NIH HHS/United States
- R37 DK047556/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous