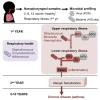
The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development - PubMed (original) (raw)
. 2015 May 13;17(5):704-15.
doi: 10.1016/j.chom.2015.03.008. Epub 2015 Apr 9.
Danny Mok 2, Kym Pham 3, Merci Kusel 2, Michael Serralha 2, Niamh Troy 2, Barbara J Holt 2, Belinda J Hales 2, Michael L Walker 4, Elysia Hollams 2, Yury A Bochkov 5, Kristine Grindle 5, Sebastian L Johnston 6, James E Gern 5, Peter D Sly 7, Patrick G Holt 8, Kathryn E Holt 9, Michael Inouye 10
Affiliations
- PMID: 25865368
- PMCID: PMC4433433
- DOI: 10.1016/j.chom.2015.03.008
The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development
Shu Mei Teo et al. Cell Host Microbe. 2015.
Abstract
The nasopharynx (NP) is a reservoir for microbes associated with acute respiratory infections (ARIs). Lung inflammation resulting from ARIs during infancy is linked to asthma development. We examined the NP microbiome during the critical first year of life in a prospective cohort of 234 children, capturing both the viral and bacterial communities and documenting all incidents of ARIs. Most infants were initially colonized with Staphylococcus or Corynebacterium before stable colonization with Alloiococcus or Moraxella. Transient incursions of Streptococcus, Moraxella, or Haemophilus marked virus-associated ARIs. Our data identify the NP microbiome as a determinant for infection spread to the lower airways, severity of accompanying inflammatory symptoms, and risk for future asthma development. Early asymptomatic colonization with Streptococcus was a strong asthma predictor, and antibiotic usage disrupted asymptomatic colonization patterns. In the absence of effective anti-viral therapies, targeting pathogenic bacteria within the NP microbiome could represent a prophylactic approach to asthma.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
Graphical abstract
Figure 1
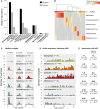
Bacterial Composition of 1,021 Nasopharyngeal Aspirates Collected from 234 Infants during Periods of Respiratory Health and Disease (A) Frequency of the most abundant phyla and genera (comprising 99.9% of reads). (B) Clustering of samples into microbiome profile groups (MPGs) based on relative abundance of the six most common genera. Colored bars indicate MPGs, labeled by their dominant genus: Moraxella (red), Corynebacterium (blue), Alloiococcus (green), Staphylococcus (purple), Haemophilus (yellow), and Streptococcus (orange). (C) Weekly frequencies of MPGs among healthy samples, collected during planned visits at approximately 2, 6, and 12 months of age and following at least 4 weeks without symptoms of acute respiratory infection (ARI). (D) Weekly frequencies of each MPG among ARI samples. (E) Odds ratios for association of MPGs with ARI symptoms, adjusted for age, gender, season, number of prior infections, antibiotics intake, mother’s antibiotics intake, delivery mode, and breastfeeding; with and without adjustment for detection of common viruses (RSV, HRV).
Figure 2
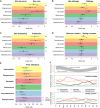
Impact of Environmental Factors on Relative Abundances of Major Genera of the NP Microbiome (A–D) Squares, odds ratios; filled squares, healthy samples; empty squares, infection samples; bars, 95% confidence intervals; ∗p < 0.1, ∗∗p < 0.05, ∗∗∗p < 0.01. Associations are estimated using logistic regression and adjusted for age: (A) day care attendance (yes versus no, 12-month samples), (B) co-habiting with siblings, (C) antibiotics intake in the 4 weeks preceding NP sample collection, (D) season (spring-summer versus autumn-winter). (E) Impact of prior ARI, estimated using proportional odds ordinal logistic regression and categorized as 0, 1, or ≥ 2 ARI. (F) Seasonal patterns: top, mean maximum and minimum temperatures in study location (Perth); bottom, monthly proportions of samples in each microbiome profile group (MPG) for infection and healthy samples.
Figure 3
Symptoms of Lower Respiratory Illness during the First Year of Life Are Associated with Viruses Present during the Infection and Predict Chronic Wheeze at 5 Years of Age (A and B) Kaplan-Meier survival curves for age (days) at (A) first febrile LRI and (B) first HRV-C wheezy LRI, stratified by chronic wheeze status at 5 years. p values shown were estimated using Cox proportional hazards models, adjusted for gender and maternal and paternal history of atopic disease. Shaded areas indicate 95% confidence intervals. (C) Frequencies of fever and wheeze symptoms during LRI, in which HRV-C and/or RSV were detected. Total numbers are: HRV-C only, n = 79; HRV-C and RSV, n = 14; RSV only, n = 22. (D) Cross-tabulation of individuals according to their experience of LRI during infancy; percentages in brackets indicate frequency of chronic wheeze at 5 years.
Figure 4
Impact of Early Colonization on Age at First Respiratory Infection (A) Microbiome profile group (MPG) transitions between healthy samples (T1) and the next sequenced infection (T2). Cell numbers indicate the number of times the respective transition from T1 to T2 was observed in the dataset; cells are colored to indicate the row proportions as per legend. (B–D) Kaplan-Meier survival curves for age (days) of first (B) acute respiratory illness (ARI), (C) upper respiratory illness (URI), and (D) lower respiratory illness (LRI), stratified according to the MPG of the first healthy sample (collected by 9 weeks of age and prior to any infection, n = 160). Cox proportional hazards models were adjusted for age, gender, season, virus status in the early healthy sample, and virus status at the first event. Shaded areas indicate 95% confidence intervals.
Figure 5
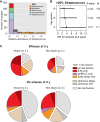
Predictors of Chronic Wheeze at Age 5 (A) Streptococcus abundance among healthy samples collected by 9 weeks of age, broken down by microbiome profile group (MPG). (B) Adjusted odds ratio (OR, squares) and 95% confidence intervals (bars) for association between chronic wheeze at age 5 and high (> 20%) abundance of Streptococcus in the first healthy NP sample; p values and sample sizes (n) are indicated; individuals who experienced an infection prior to first healthy NP sample collection were excluded. (C) Distribution of microbial events during infancy that were identified as risk factors for chronic wheeze at 5 years of age, stratified according to atopic status by age 2. fLRI, febrile LRI; wHRV-C, HRV-C LRI accompanied by wheeze; Strep, > 20% Streptococcus abundance in healthy NP sample taken in by 9 weeks old and prior to any ARI; unknown Strep, no such NP sample available (mainly due to ARI before 9 weeks of age); unknown LRI, incomplete viral/symptom profiling for LRI. Size of pie chart is proportional to the number of infants in each condition.
Comment in
- Infectious asthma triggers: time to revise the hygiene hypothesis?
Webley WC, Aldridge KL. Webley WC, et al. Trends Microbiol. 2015 Jul;23(7):389-91. doi: 10.1016/j.tim.2015.05.006. Epub 2015 Jun 9. Trends Microbiol. 2015. PMID: 26070971
Similar articles
- The nasopharyngeal microbiota in patients with viral respiratory tract infections is enriched in bacterial pathogens.
Edouard S, Million M, Bachar D, Dubourg G, Michelle C, Ninove L, Charrel R, Raoult D. Edouard S, et al. Eur J Clin Microbiol Infect Dis. 2018 Sep;37(9):1725-1733. doi: 10.1007/s10096-018-3305-8. Epub 2018 Jul 22. Eur J Clin Microbiol Infect Dis. 2018. PMID: 30033505 - Associations Between Viral and Bacterial Potential Pathogens in the Nasopharynx of Children With and Without Respiratory Symptoms.
Skevaki CL, Tsialta P, Trochoutsou AI, Logotheti I, Makrinioti H, Taka S, Lebessi E, Paraskakis I, Papadopoulos NG, Tsolia MN. Skevaki CL, et al. Pediatr Infect Dis J. 2015 Dec;34(12):1296-301. doi: 10.1097/INF.0000000000000872. Pediatr Infect Dis J. 2015. PMID: 26262821 - Airway Microbiota Dynamics Uncover a Critical Window for Interplay of Pathogenic Bacteria and Allergy in Childhood Respiratory Disease.
Teo SM, Tang HHF, Mok D, Judd LM, Watts SC, Pham K, Holt BJ, Kusel M, Serralha M, Troy N, Bochkov YA, Grindle K, Lemanske RF Jr, Johnston SL, Gern JE, Sly PD, Holt PG, Holt KE, Inouye M. Teo SM, et al. Cell Host Microbe. 2018 Sep 12;24(3):341-352.e5. doi: 10.1016/j.chom.2018.08.005. Cell Host Microbe. 2018. PMID: 30212648 Free PMC article. - Relationship between nasopharyngeal microbiota and patient's susceptibility to viral infection.
Dubourg G, Edouard S, Raoult D. Dubourg G, et al. Expert Rev Anti Infect Ther. 2019 Jun;17(6):437-447. doi: 10.1080/14787210.2019.1621168. Epub 2019 May 29. Expert Rev Anti Infect Ther. 2019. PMID: 31106653 Review. - Nasal Microbiome and Its Interaction with the Host in Childhood Asthma.
Zeng Y, Liang JQ. Zeng Y, et al. Cells. 2022 Oct 7;11(19):3155. doi: 10.3390/cells11193155. Cells. 2022. PMID: 36231116 Free PMC article. Review.
Cited by
- A complete guide to human microbiomes: Body niches, transmission, development, dysbiosis, and restoration.
Reynoso-García J, Miranda-Santiago AE, Meléndez-Vázquez NM, Acosta-Pagán K, Sánchez-Rosado M, Díaz-Rivera J, Rosado-Quiñones AM, Acevedo-Márquez L, Cruz-Roldán L, Tosado-Rodríguez EL, Figueroa-Gispert MDM, Godoy-Vitorino F. Reynoso-García J, et al. Front Syst Biol. 2022;2:951403. doi: 10.3389/fsysb.2022.951403. Epub 2022 Jul 22. Front Syst Biol. 2022. PMID: 38993286 Free PMC article. - Microbiota profiles in pre-school children with respiratory infections: Modifications induced by the oral bacterial lysate OM-85.
Esposito S, Ballarini S, Argentiero A, Ruggiero L, Rossi GA, Principi N. Esposito S, et al. Front Cell Infect Microbiol. 2022 Aug 16;12:789436. doi: 10.3389/fcimb.2022.789436. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36051241 Free PMC article. Clinical Trial. - Respiratory syncytial virus and rhinovirus severe bronchiolitis are associated with distinct nasopharyngeal microbiota.
Mansbach JM, Hasegawa K, Henke DM, Ajami NJ, Petrosino JF, Shaw CA, Piedra PA, Sullivan AF, Espinola JA, Camargo CA Jr. Mansbach JM, et al. J Allergy Clin Immunol. 2016 Jun;137(6):1909-1913.e4. doi: 10.1016/j.jaci.2016.01.036. Epub 2016 Apr 6. J Allergy Clin Immunol. 2016. PMID: 27061249 Free PMC article. Clinical Trial. No abstract available. - Winds of change a tale of: asthma and microbiome.
Galeana-Cadena D, Gómez-García IA, Lopez-Salinas KG, Irineo-Moreno V, Jiménez-Juárez F, Tapia-García AR, Boyzo-Cortes CA, Matías-Martínez MB, Jiménez-Alvarez L, Zúñiga J, Camarena A. Galeana-Cadena D, et al. Front Microbiol. 2023 Dec 11;14:1295215. doi: 10.3389/fmicb.2023.1295215. eCollection 2023. Front Microbiol. 2023. PMID: 38146448 Free PMC article. Review. - Oral Cavity Microbiome Impact on Respiratory Infections Among Children.
Crestez AM, Nechita A, Daineanu MP, Busila C, Tatu AL, Ionescu MA, Martinez JD, Debita M. Crestez AM, et al. Pediatric Health Med Ther. 2024 Oct 9;15:311-323. doi: 10.2147/PHMT.S471588. eCollection 2024. Pediatric Health Med Ther. 2024. PMID: 39398897 Free PMC article. Review.
References
- Arrieta M.C., Finlay B. The intestinal microbiota and allergic asthma. J. Infect. 2014;69(1):S53–S55. - PubMed
- Bisgaard H., Hermansen M.N., Buchvald F., Loland L., Halkjaer L.B., Bønnelykke K., Brasholt M., Heltberg A., Vissing N.H., Thorsen S.V. Childhood asthma after bacterial colonization of the airway in neonates. N. Engl. J. Med. 2007;357:1487–1495. - PubMed
Publication types
MeSH terms
Grants and funding
- P01 HL070831/HL/NHLBI NIH HHS/United States
- U19 AI070503/AI/NIAID NIH HHS/United States
- U19 AI104317/AI/NIAID NIH HHS/United States
- G1000758/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous