Metabolic Dependencies in RAS-Driven Cancers - PubMed (original) (raw)
Review
Metabolic Dependencies in RAS-Driven Cancers
Alec C Kimmelman. Clin Cancer Res. 2015.
Abstract
The ability to inhibit the RAS oncogene has been the holy grail of oncology because of the critical role of this gene in a multitude of tumor types. In addition, RAS-mutant tumors are among the most aggressive and refractory to treatment. Although directly targeting the RAS oncogene has proven challenging, an alternative approach for treating RAS-driven cancers is to inhibit critical downstream events that are required for tumor maintenance. Indeed, much focus has been put on inhibiting signaling cascades downstream of RAS. Recent studies have shown that oncogenic RAS promotes a metabolic reprogramming of tumor cells, shifting them toward an anabolic metabolism necessary to produce biomass to support unconstrained proliferation. These cancers also use a diverse set of fuel sources to meet their metabolic needs and have even developed a variety of mechanisms to act as metabolic scavengers to obtain necessary metabolic substrates from both extracellular and intracellular sources. Collectively, these adaptations can create "metabolic bottlenecks" whereby tumor cells rely on particular pathways or rate-limiting metabolites. In this regard, inhibiting individual or combinations of these metabolic pathways can attenuate growth in preclinical models. Because these dependencies are tumor selective and downstream of oncogenic RAS, there is the opportunity for therapeutic intervention. Although targeting tumor metabolism is still in the early days of translation to patients, our continued advances in understanding critical metabolic adaptations in RAS-driven cancers, as well as the ability to study this altered metabolism in relevant tumor models, will accelerate the development of new therapeutic approaches. Clin Cancer Res; 21(8); 1828-34. ©2015 AACR. See all articles in this CCR Focus section, "Targeting RAS-Driven Cancers."
©2015 American Association for Cancer Research.
Conflict of interest statement
Disclosure of Potential Conflicts of Interest
A.C. Kimmelman reports receiving speakers bureau honoraria from Agios and the US Oncology Network, and is a consultant/advisory board member for Astellas Pharma, FORMA Therapeutics, and Gilead. No other potential conflicts of interest were disclosed.
Figures
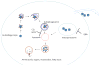
Figure 1
Metabolic scavenging pathways have critical roles in Ras-driven cancers. Autophagy can degrade intracellular cargo by sequestering it in autophagosomes. These fuse with lysosomes where the cargo is degraded and subsequently recycled back into the cytosol. Macropinocytosis is associated with membrane ruffling that can lead to the internalization of extracellular material such as albumin with subsequent lysosomal degradation. Both pathways can supply metabolic substrates for various metabolic pathways. Chloroquine (CQ) and hydroxychloroquine (HCQ) inhibit lysosomal acidification and therefore block the degradation of cargo in both pathways. Ethylisopropylamiloride (EIPA) inhibits macropinocytosis.
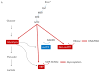
Figure 2
Oncogenic Kras (KRAS*) can create metabolic alterations in pancreatic tumor cells. (A). Activation of the of the RAF/MEK/ERK cascade leads to increased MYC levels which promotes increased expression of various enzymes involved in glucose metabolism (representative enzymes that are increased are indicated). This promotes increased glycolysis as well as increased flux of glucose derived carbon through anabolic pathways such as the non-oxidative arm of the pentose phosphate pathway (PPP) to produce ribose for DNA/RNA biosynthesis and the hexosamine biosynthesis pathway (HBP) to produce glycosylation precursors. Kras regulated pathways are depicted in red and representative enzymes upregulated by oncogenic Kras are indicated for each pathway. The oxidative PPP (in purple) which can produce NADPH is not regulated by Kras. OAA (oxaloacetate); Asp (aspartate) (B). Kras causes a shift in glutamine metabolism in pancreatic cancers, through an unknown downstream program, repressing GLUD1 and increasing GOT1 expression. This pathway is a major source of cytosolic NADPH in Kras mutant pancreatic cancer cells and is critical for redox balance. Both mitochondrial (the structure depicted is a mitochondria) and cytosolic enzymes are involved in this pathway.
Figure 2
Oncogenic Kras (KRAS*) can create metabolic alterations in pancreatic tumor cells. (A). Activation of the of the RAF/MEK/ERK cascade leads to increased MYC levels which promotes increased expression of various enzymes involved in glucose metabolism (representative enzymes that are increased are indicated). This promotes increased glycolysis as well as increased flux of glucose derived carbon through anabolic pathways such as the non-oxidative arm of the pentose phosphate pathway (PPP) to produce ribose for DNA/RNA biosynthesis and the hexosamine biosynthesis pathway (HBP) to produce glycosylation precursors. Kras regulated pathways are depicted in red and representative enzymes upregulated by oncogenic Kras are indicated for each pathway. The oxidative PPP (in purple) which can produce NADPH is not regulated by Kras. OAA (oxaloacetate); Asp (aspartate) (B). Kras causes a shift in glutamine metabolism in pancreatic cancers, through an unknown downstream program, repressing GLUD1 and increasing GOT1 expression. This pathway is a major source of cytosolic NADPH in Kras mutant pancreatic cancer cells and is critical for redox balance. Both mitochondrial (the structure depicted is a mitochondria) and cytosolic enzymes are involved in this pathway.
Similar articles
- The greedy nature of mutant RAS: a boon for drug discovery targeting cancer metabolism?
Lv J, Wang J, Chang S, Liu M, Pang X. Lv J, et al. Acta Biochim Biophys Sin (Shanghai). 2016 Jan;48(1):17-26. doi: 10.1093/abbs/gmv102. Epub 2015 Oct 19. Acta Biochim Biophys Sin (Shanghai). 2016. PMID: 26487443 Review. - Molecular pathways: targeting the dependence of mutant RAS cancers on the DNA damage response.
Grabocka E, Commisso C, Bar-Sagi D. Grabocka E, et al. Clin Cancer Res. 2015 Mar 15;21(6):1243-7. doi: 10.1158/1078-0432.CCR-14-0650. Epub 2014 Nov 25. Clin Cancer Res. 2015. PMID: 25424849 Free PMC article. Review. - MTH1 expression is required for effective transformation by oncogenic HRAS.
Giribaldi MG, Munoz A, Halvorsen K, Patel A, Rai P. Giribaldi MG, et al. Oncotarget. 2015 May 10;6(13):11519-29. doi: 10.18632/oncotarget.3447. Oncotarget. 2015. PMID: 25893378 Free PMC article. - Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth.
Gaglio D, Metallo CM, Gameiro PA, Hiller K, Danna LS, Balestrieri C, Alberghina L, Stephanopoulos G, Chiaradonna F. Gaglio D, et al. Mol Syst Biol. 2011 Aug 16;7:523. doi: 10.1038/msb.2011.56. Mol Syst Biol. 2011. PMID: 21847114 Free PMC article. - Evaluating The Role Of Nitric Oxide Synthase In Oncogenic Ras-Driven Tumorigenesis.
Counter C. Counter C. Redox Biol. 2015 Aug;5:417. doi: 10.1016/j.redox.2015.09.023. Epub 2015 Dec 30. Redox Biol. 2015. PMID: 28162280
Cited by
- Targeting the Metabolic Adaptation of Metastatic Cancer.
Tarragó-Celada J, Cascante M. Tarragó-Celada J, et al. Cancers (Basel). 2021 Apr 1;13(7):1641. doi: 10.3390/cancers13071641. Cancers (Basel). 2021. PMID: 33915900 Free PMC article. Review. - Macropinocytosis Exploitation by Cancers and Cancer Therapeutics.
Ha KD, Bidlingmaier SM, Liu B. Ha KD, et al. Front Physiol. 2016 Sep 12;7:381. doi: 10.3389/fphys.2016.00381. eCollection 2016. Front Physiol. 2016. PMID: 27672367 Free PMC article. Review. - Oncogenic Mechanisms and Therapeutic Targeting of Metabolism in Leukemia and Lymphoma.
Stahl M, Epstein-Peterson ZD, Intlekofer AM. Stahl M, et al. Cold Spring Harb Perspect Med. 2021 Jul 1;11(7):a035477. doi: 10.1101/cshperspect.a035477. Cold Spring Harb Perspect Med. 2021. PMID: 32816875 Free PMC article. Review. - Oncogenic KRAS triggers metabolic reprogramming in pancreatic ductal adenocarcinoma.
Shen X, Niu N, Xue J. Shen X, et al. J Transl Int Med. 2022 Nov 15;11(4):322-329. doi: 10.2478/jtim-2022-0022. eCollection 2023 Dec. J Transl Int Med. 2022. PMID: 38130635 Free PMC article. - Defining a metabolic landscape of tumours: genome meets metabolism.
Seth Nanda C, Venkateswaran SV, Patani N, Yuneva M. Seth Nanda C, et al. Br J Cancer. 2020 Jan;122(2):136-149. doi: 10.1038/s41416-019-0663-7. Epub 2019 Dec 10. Br J Cancer. 2020. PMID: 31819196 Free PMC article. Review.
References
- Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25:272–81. - PubMed
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources