The dnd operon for DNA phosphorothioation modification system in Escherichia coli is located in diverse genomic islands - PubMed (original) (raw)
The dnd operon for DNA phosphorothioation modification system in Escherichia coli is located in diverse genomic islands
Wing Sze Ho et al. BMC Genomics. 2015.
Abstract
Background: Strains of Escherichia coli that are non-typeable by pulsed-field gel electrophoresis (PFGE) due to in-gel degradation can influence their molecular epidemiological data. The DNA degradation phenotype (Dnd(+)) is mediated by the dnd operon that encode enzymes catalyzing the phosphorothioation of DNA, rendering the modified DNA susceptible to oxidative cleavage during a PFGE run. In this study, a PCR assay was developed to detect the presence of the dnd operon in Dnd(+) E. coli strains and to improve their typeability. Investigations into the genetic environments of the dnd operon in various E. coli strains led to the discovery that the dnd operon is harboured in various diverse genomic islands.
Results: The dndBCDE genes (dnd operon) were detected in all Dnd(+) E. coli strains by PCR. The addition of thiourea improved the typeability of Dnd(+) E. coli strains to 100% using PFGE and the Dnd(+) phenotype can be observed in both clonal and genetically diverse E. coli strains. Genomic analysis of 101 dnd operons from genome sequences of Enterobacteriaceae revealed that the dnd operons of the same bacterial species were generally clustered together in the phylogenetic tree. Further analysis of dnd operons of 52 E. coli genomes together with their respective immediate genetic environments revealed a total of 7 types of genetic organizations, all of which were found to be associated with genomic islands designated dnd-encoding GIs. The dnd-encoding GIs displayed mosaic structure and the genomic context of the 7 islands (with 1 representative genome from each type of genetic organization) were also highly variable, suggesting multiple recombination events. This is also the first report where two dnd operons were found within a strain although the biological implication is unknown. Surprisingly, dnd operons were frequently found in pathogenic E. coli although their link with virulence has not been explored.
Conclusion: Genomic islands likely play an important role in facilitating the horizontal gene transfer of the dnd operons in E. coli with 7 different types of islands discovered so far.
Figures
Figure 1
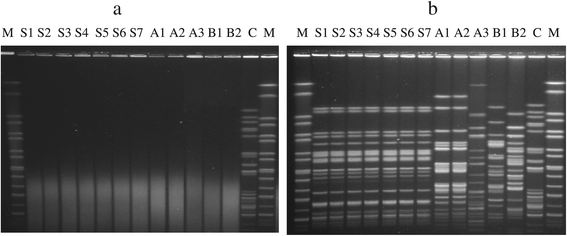
Effects of thiourea improved the typeability of Dnd + strains by PFGE. a: PFGE gel obtained by performing PFGE run without the addition of thiourea. b: PFGE gel obtained by performing PFGE run with the addition of thiourea to the agarose gel and running buffer. Lane M: XbaI digested H9812 DNA marker; lanes S1-S7, VTEC isolates from pigs; lanes A1-A3, clinical isolates obtained from medical center A; lanes B1-B2, clinical isolates obtained from medical center B; C, control E. coli isolate which is typeable with and without the addition of thiourea (a and b). Gel images were captured using GelDoc (BioRad, Hercules, CA) digital gel documentation system.
Figure 2
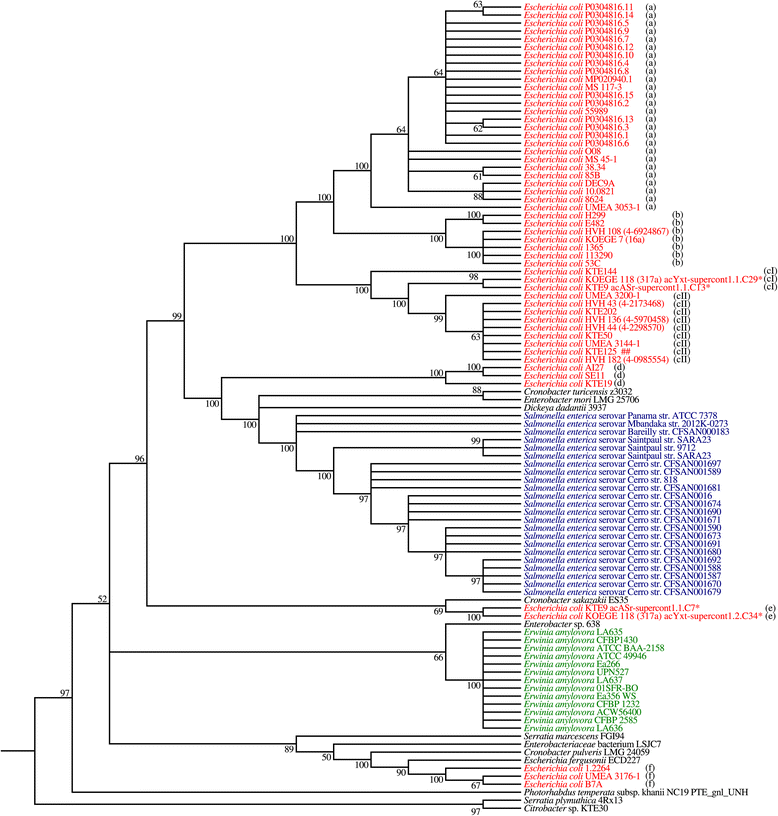
Phylogenetic tree of dnd operons from Enterobacteriaceae . Asterisk (*) indicates that the genome contains two copies of dnd operons. Different alphabets (a, b, cI, cII, d, e, f) depict the subgroups based on the immediate genetic environment. Colored genome strain codes facilitate visualization. Maximum likelihood (ML) method was used to construct the phylogenetic tree using MEGA5. Bootstrap confidence values greater than 50% are shown at branches. Nodes with less than 50% bootstrap value are collapsed.
Figure 3
Genetic map of 7 different dnd- encoding genomic islands of E . coli . Same-strand DNA similarity is shaded blue (dark blue-sky blue) while reverse similarity is shaded red (red-light pink). Coding sequences are displayed as arrows. Major features are displayed and are colored to facilitate visualization. Order of genomic islands in the map was arranged according to the phylogenetic tree constructed using the dnd operons. Red alphabet in brackets indicates the subgroup determined based on the immediate genetic environment of the respective dnd operons. Integrase gene is the first element of the _dnd_-encoding genomic island (GI).
Figure 4
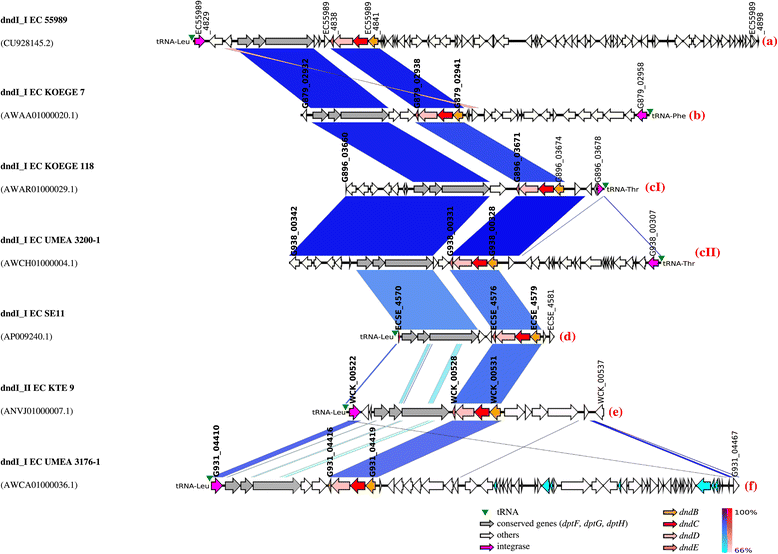
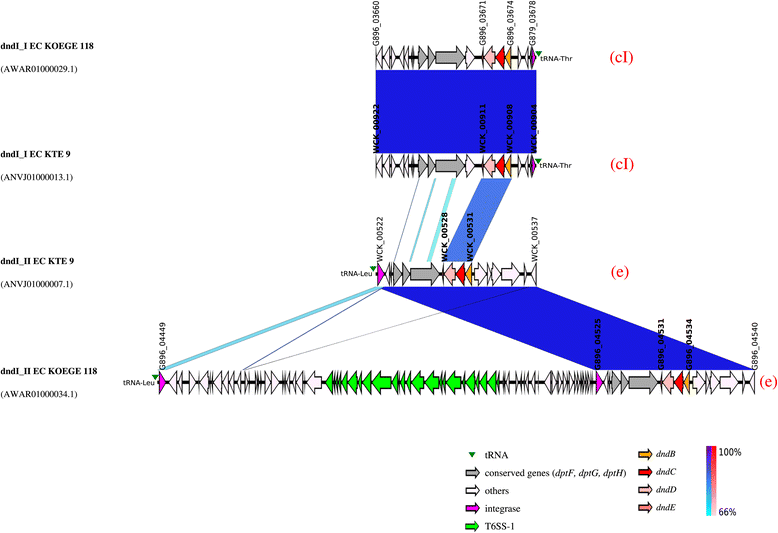
Genetic map of the dnd- encoding genomic islands of E . coli KTE9 and KOEGE118 which harboured 2 copies of dnd operons. The integrase gene is the first element of the _dnd_-encoding genomic island (GI) although the genetic map is illustrated according to the orientation of the dnd operons to facilitate visualization. Same-strand DNA similarity is shaded blue (dark blue-sky blue) while reverse similarity is shaded red (red-light pink). Coding sequences are displayed as arrows. Major features are displayed and are colored to facilitate visualization. Red alphabet in brackets indicates the subgroup determined based on the immediate genetic environment of the respective dnd operons. An additional DNA segment of 74 kb was observed for dndI_IIEC KOEGE 118 compared to dndI_IIEC KTE 9. dndI_IEC KOEGE 118 is identical to dndI_IEC KTE 9.
Figure 5
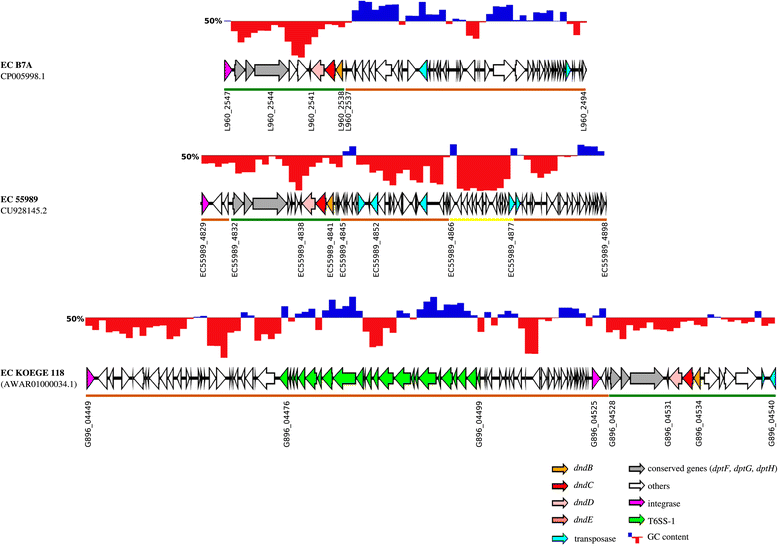
GC content of three representative dnd -encoding genomic islands. GC content of _dnd_-encoding GIs are shown above their respective genetic maps, with blue and red region showing GC content above and below 50%, respectively. The _dnd_-encoding GIs consist of several modules, depicted by the coloured bars below their respective genetic maps. ORFs above the brown coloured bars were found in both _dnd_-positive and _dnd_-negative E. coli genomes. ORFs that are underlined by green bars are strictly associated with _dnd_-positive genomes only while ORFs underlined by the yellow bar are strain-specific.
Figure 6
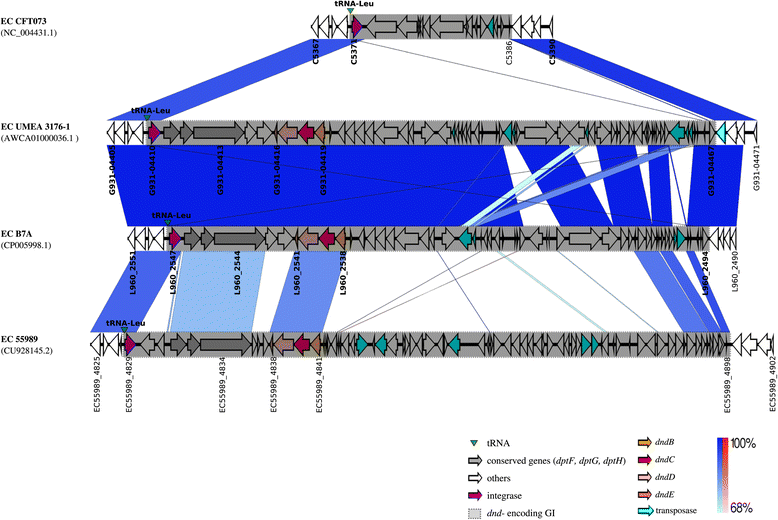
Example of different genetic contents at the leu tRNA insertion sites of dnd -negative and dnd -positive E . coli genomes. Same-strand DNA similarity is shaded blue (dark blue-sky blue) while reverse similarity is shaded red (red-light pink). Coding sequences are displayed as arrows. Major features are displayed and are coloured to facilitate visualization.
Similar articles
- Insertion site and distribution of a genomic island conferring DNA phosphorothioation in the Mycobacterium abscessus complex.
Howard ST, Newman KL, McNulty S, Brown-Elliott BA, Vasireddy R, Bridge L, Wallace RJ. Howard ST, et al. Microbiology (Reading). 2013 Nov;159(Pt 11):2323-2332. doi: 10.1099/mic.0.070318-0. Epub 2013 Sep 6. Microbiology (Reading). 2013. PMID: 24014661 - From the Flavobacterium genus to the phylum Bacteroidetes: genomic analysis of dnd gene clusters.
Barbier P, Lunazzi A, Fujiwara-Nagata E, Avendaño-Herrera R, Bernardet JF, Touchon M, Duchaud E. Barbier P, et al. FEMS Microbiol Lett. 2013 Nov;348(1):26-35. doi: 10.1111/1574-6968.12239. Epub 2013 Sep 6. FEMS Microbiol Lett. 2013. PMID: 23965156 - DNA looping in prokaryotes: experimental and theoretical approaches.
Cournac A, Plumbridge J. Cournac A, et al. J Bacteriol. 2013 Mar;195(6):1109-19. doi: 10.1128/JB.02038-12. Epub 2013 Jan 4. J Bacteriol. 2013. PMID: 23292776 Free PMC article. Review. - Twenty years hunting for sulfur in DNA.
Chen S, Wang L, Deng Z. Chen S, et al. Protein Cell. 2010 Jan;1(1):14-21. doi: 10.1007/s13238-010-0009-y. Epub 2010 Feb 7. Protein Cell. 2010. PMID: 21203994 Free PMC article. Review.
Cited by
- New Insights into the Genetic Diversity of Clostridium botulinum Group III through Extensive Genome Exploration.
Woudstra C, Le Maréchal C, Souillard R, Bayon-Auboyer MH, Mermoud I, Desoutter D, Fach P. Woudstra C, et al. Front Microbiol. 2016 May 19;7:757. doi: 10.3389/fmicb.2016.00757. eCollection 2016. Front Microbiol. 2016. PMID: 27242769 Free PMC article. - Structural investigation into physiological DNA phosphorothioate modification.
Lan W, Hu Z, Shen J, Wang C, Jiang F, Liu H, Long D, Liu M, Cao C. Lan W, et al. Sci Rep. 2016 May 12;6:25737. doi: 10.1038/srep25737. Sci Rep. 2016. PMID: 27169778 Free PMC article. - DNA Phosphorothioate Modifications Are Widely Distributed in the Human Microbiome.
Sun Y, Kong L, Wu G, Cao B, Pang X, Deng Z, Dedon PC, Zhang C, You D. Sun Y, et al. Biomolecules. 2020 Aug 12;10(8):1175. doi: 10.3390/biom10081175. Biomolecules. 2020. PMID: 32806589 Free PMC article. - Comparative genomics reveals structural and functional features specific to the genome of a foodborne Escherichia coli O157:H7.
Sharma VK, Akavaram S, Schaut RG, Bayles DO. Sharma VK, et al. BMC Genomics. 2019 Mar 8;20(1):196. doi: 10.1186/s12864-019-5568-6. BMC Genomics. 2019. PMID: 30849935 Free PMC article. - Serratia liquefaciens FG3 isolated from a metallophyte plant sheds light on the evolution and mechanisms of adaptive traits in extreme environments.
Caneschi WL, Sanchez AB, Felestrino ÉB, Lemes CGC, Cordeiro IF, Fonseca NP, Villa MM, Vieira IT, Moraes LÂG, Assis RAB, do Carmo FF, Kamino LHY, Silva RS, Ferro JA, Ferro MIT, Ferreira RM, Santos VL, Silva UCM, Almeida NF, Varani AM, Garcia CCM, Setubal JC, Moreira LM. Caneschi WL, et al. Sci Rep. 2019 Nov 29;9(1):18006. doi: 10.1038/s41598-019-54601-4. Sci Rep. 2019. PMID: 31784663 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases