Insights into the evolutionary history of Japanese encephalitis virus (JEV) based on whole-genome sequences comprising the five genotypes - PubMed (original) (raw)
Insights into the evolutionary history of Japanese encephalitis virus (JEV) based on whole-genome sequences comprising the five genotypes
Xiaoyan Gao et al. Virol J. 2015.
Abstract
Background: Japanese encephalitis virus (JEV) is the etiological agent of Japanese encephalitis (JE), one of the most serious viral encephalitis worldwide. Five genotypes have been classified based on phylogenetic analysis of the viral envelope gene or the complete genome. Previous studies based on four genotypes have reported that in evolutionary terms, genotype 1 JEV is the most recent lineage. However, until now, no systematic phylogenetic analysis was reported based on whole genomic sequence of all five JEV genotypes.
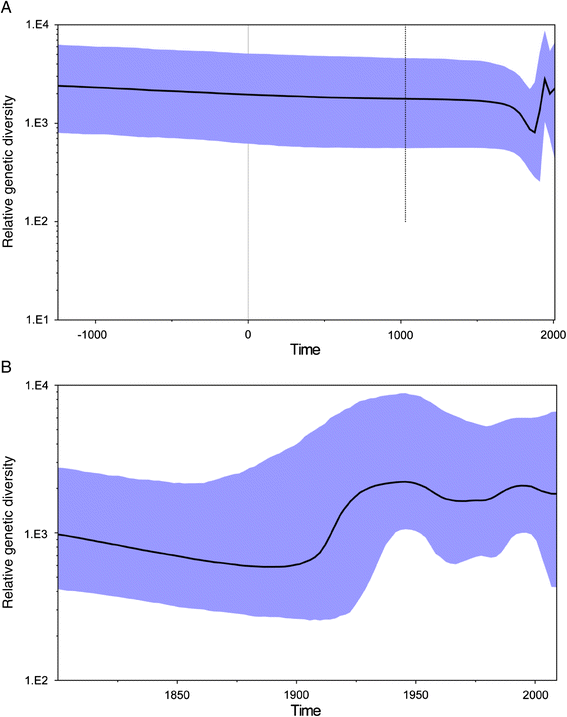
Findings: In this study, phylogenetic analysis using Bayesian Markov chain Monte Carlo simulations was conducted on the whole genomic sequences of all five genotypes of JEV. The results showed that the most recent common ancestor (TMRCA) for JEV is estimated to have occurred 3255 years ago (95% highest posterior density [HPD], -978 to-6125 years). Chronologically, this ancestral lineage diverged to produce five recognized virus genotypes in the sequence 5, 4, 3, 2 and 1. Population dynamics analysis indicated that the genetic diversity of the virus peaked during the following two periods: 1930-1960 and 1980-1990, and the population diversity of JEV remained relatively high after 2000.
Conclusions: Genotype 5 is the earliest recognized JEV lineage, and the genetic diversity of JEV has remained high since 2000.
Figures
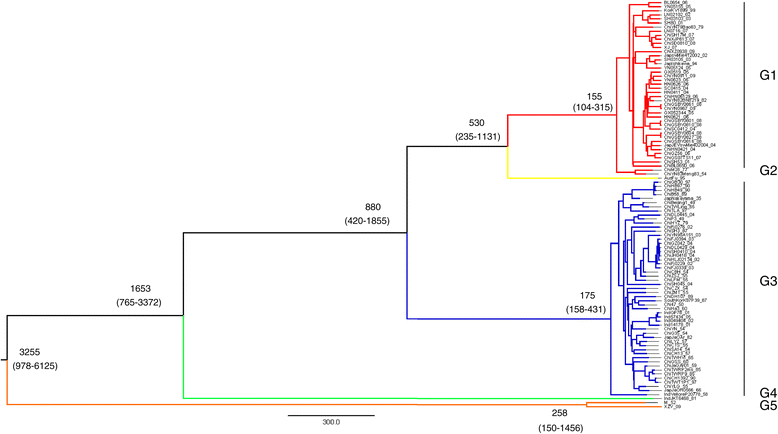
Figure 1
Maximum clade credibility (MCC) tree for 100 whole-genome sequences of JEV. Five distinct lineages were identified: G1 (red), G2 (yellow), G3 (blue), G4 (green) and G5 (orange). Estimated TMRCAs of these lineages (with their 95% HPD values in parentheses) are shown, G1: 155(104–315), G2: 530(235–1131), G3: 880(420–1855), G4: 1653(765–3372), and G5: 3255(978–6125).
Figure 2
Bayesian skyline plots for JEV. Highlighted areas correspond to 95% HPD intervals. (A) Populations during the whole evolutionary history; (B) Populations during the later evolutionary history since 1800.
Similar articles
- Molecular epidemiology of Japanese encephalitis in northern Vietnam, 1964-2011: genotype replacement.
Do LP, Bui TM, Hasebe F, Morita K, Phan NT. Do LP, et al. Virol J. 2015 Apr 1;12:51. doi: 10.1186/s12985-015-0278-4. Virol J. 2015. PMID: 25889499 Free PMC article. - Genetic diversity of Japanese encephalitis virus isolates obtained from the Indonesian archipelago between 1974 and 1987.
Schuh AJ, Guzman H, Tesh RB, Barrett AD. Schuh AJ, et al. Vector Borne Zoonotic Dis. 2013 Jul;13(7):479-88. doi: 10.1089/vbz.2011.0870. Epub 2013 Apr 16. Vector Borne Zoonotic Dis. 2013. PMID: 23590316 Free PMC article. - [Molecular characteristics of the genome of G I of Japanese encephalitis virus isolated from the specimen collected from viral encephalitis case for the first time].
Li J, Fu SH, Wang LH, Gao XY, Wang HY, Ye XF, Zhao SY, Liu CT, Zhu WY, Wang L, Liang GD. Li J, et al. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2012 Apr;26(2):84-6. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2012. PMID: 23002539 Chinese. - Japanese encephalitis and Japanese encephalitis virus in mainland China.
Zheng Y, Li M, Wang H, Liang G. Zheng Y, et al. Rev Med Virol. 2012 Sep;22(5):301-22. doi: 10.1002/rmv.1710. Epub 2012 Mar 8. Rev Med Virol. 2012. PMID: 22407526 Review. - The reemerging and outbreak of genotypes 4 and 5 of Japanese encephalitis virus.
Zhang W, Yin Q, Wang H, Liang G. Zhang W, et al. Front Cell Infect Microbiol. 2023 Nov 16;13:1292693. doi: 10.3389/fcimb.2023.1292693. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 38076463 Free PMC article. Review.
Cited by
- Adaptive Evolution as a Driving Force of the Emergence and Re-Emergence of Mosquito-Borne Viral Diseases.
Yu X, Cheng G. Yu X, et al. Viruses. 2022 Feb 21;14(2):435. doi: 10.3390/v14020435. Viruses. 2022. PMID: 35216028 Free PMC article. Review. - Japanese Encephalitis Virus: An Update on the Potential Antivirals and Vaccines.
Srivastava KS, Jeswani V, Pal N, Bohra B, Vishwakarma V, Bapat AA, Patnaik YP, Khanna N, Shukla R. Srivastava KS, et al. Vaccines (Basel). 2023 Mar 27;11(4):742. doi: 10.3390/vaccines11040742. Vaccines (Basel). 2023. PMID: 37112654 Free PMC article. Review. - Peripheral Nerve Injury Induced by Japanese Encephalitis Virus in C57BL/6 Mouse.
Yang H, Wang X, Wang Z, Wang G, Fu S, Li F, Yang L, Yuan Y, Shen K, Wang H, Wang Z. Yang H, et al. J Virol. 2023 May 31;97(5):e0165822. doi: 10.1128/jvi.01658-22. Epub 2023 Apr 18. J Virol. 2023. PMID: 37071015 Free PMC article. - Beyond the Surface: Endocytosis of Mosquito-Borne Flaviviruses.
Carro SD, Cherry S. Carro SD, et al. Viruses. 2020 Dec 23;13(1):13. doi: 10.3390/v13010013. Viruses. 2020. PMID: 33374822 Free PMC article. Review. - Serological and molecular epidemiology of Japanese Encephalitis in Zhejiang, China, 2015-2018.
Deng X, Yan JY, He HQ, Yan R, Sun Y, Tang XW, Zhou Y, Pan JH, Mao HY, Zhang YJ, Lv HK. Deng X, et al. PLoS Negl Trop Dis. 2020 Aug 27;14(8):e0008574. doi: 10.1371/journal.pntd.0008574. eCollection 2020 Aug. PLoS Negl Trop Dis. 2020. PMID: 32853274 Free PMC article.
References
- Uchil PD, Satchidanandam V. Phylogenetic analysis of Japanese encephalitis virus: envelope gene based analysis reveals a fifth genotype, geographic clustering, and multiple introductions of the virus into the Indian subcontinent. Am J Trop Med Hyg. 2001;65(3):242–51. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources