CopywriteR: DNA copy number detection from off-target sequence data - PubMed (original) (raw)
doi: 10.1186/s13059-015-0617-1.
Arno Velds 2, Kristel Kemper 3, Marco Ranzani 4, Lorenzo Bombardelli 5, Marlous Hoogstraat 6, Ekaterina Nevedomskaya 7 8, Guotai Xu 9, Julian de Ruiter 10 11, Martijn P Lolkema 12, Bauke Ylstra 13, Jos Jonkers 14, Sven Rottenberg 15 16, Lodewyk F Wessels 17, David J Adams 18, Daniel S Peeper 19, Oscar Krijgsman 20
Affiliations
- PMID: 25887352
- PMCID: PMC4396974
- DOI: 10.1186/s13059-015-0617-1
CopywriteR: DNA copy number detection from off-target sequence data
Thomas Kuilman et al. Genome Biol. 2015.
Abstract
Current methods for detection of copy number variants (CNV) and aberrations (CNA) from targeted sequencing data are based on the depth of coverage of captured exons. Accurate CNA determination is complicated by uneven genomic distribution and non-uniform capture efficiency of targeted exons. Here we present CopywriteR, which eludes these problems by exploiting 'off-target' sequence reads. CopywriteR allows for extracting uniformly distributed copy number information, can be used without reference, and can be applied to sequencing data obtained from various techniques including chromatin immunoprecipitation and target enrichment on small gene panels. CopywriteR outperforms existing methods and constitutes a widely applicable alternative to available tools.
Figures
Figure 1
Copy number information can be obtained from off-target reads. (A) Screenshot from the IGV genome browser, showing an example of a genomic region with sequence reads mapping to the genome before and after removal of reads in Model-based Analysis for ChIPseq (MACS) called peaks. In addition, the location of MACS-peaks, capture regions, and genes are shown. (B) Germline DNA sample C41 was subjected to WES with capture set Agilent SureSelect Human Exon Kit V4. The nature of MACS-peaks that do not overlap capture regions is displayed. The fraction of these orphan peaks that overlap with pseudogene or Ensembl exons, that do not map to any of the reference genome chromosomes, that are unmappable, and that do not belong to any of these categories are shown. (C) The distribution of sequence reads of both germline and tumor DNA samples is shown for the indicated capture sets. Sequence reads are classified into one of these categories: (1) low mapping quality reads (Phred-score < 37 and/or reads do not pair properly); (2) mitochondrial reads; (3) reads in MACS-peaks; (4) remaining reads. Error bars represent standard deviations. (D) Germline DNA sample C45 was subjected to WES, and the amount of reads after compensation for reduced effective bin size is calculated and compared to the corresponding read counts from an exon-based method. Density plots of the number of sequence reads per data point are shown for each method. (E) A flowchart of the steps incorporated in the CopywriteR tool.
Figure 2
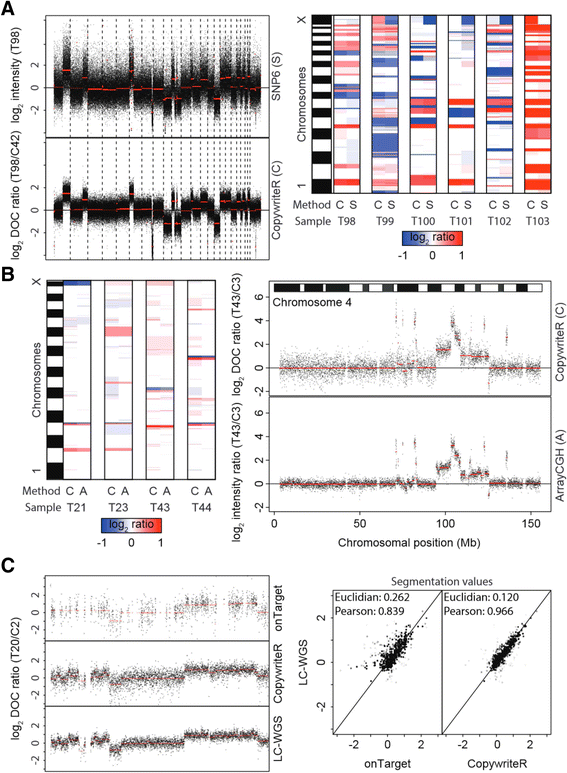
CopywriteR compares to dedicated copy number detection methods. (A) Six PDX-derived human melanoma were subjected to WES and analyzed on SNP6 arrays. Pseudo counts were derived (see Materials and methods), and used as a basis for copy number profiles, with segmentation values (CBS) depicted in red (left panel). After segmentation, segmentation values were represented as a heatmap to show concordance of the two methods. (B) Four murine small-cell lung carcinomas (SCLC) were subjected to WES and analyzed by arrayCGH. Pseudo counts were created and used for creating copy number profiles, with segmentation values (CBS) depicted in red (right panel). Segmentation values were plotted as in (A) for comparison of the two methods (left panel). (C) Tumor T20 from a breast cancer mouse model was subjected to WES or LC-WGS. Copy number profiles of chromosome 12 generated with onTarget or CopywriteR methods are compared to the profile from LC-WGS data of the same material, with segmentation values (CBS) depicted in red (left panel). Segmentation values of onTarget and CopywriteR methods are plotted against the LC-WGS method, and Euclidian distances and Pearson correlation coefficients of segmentation values are displayed (right panel).
Figure 3
CopywriteR outperforms exonic depth of coverage-based methods. (A) Tumors from a breast cancer mouse model were subjected to WES or LC-WGS, and analyzed using CopywriteR or onTarget methods. Subsequently, copy number data were segmented using propSeg or CBS, while the integrated EXCAVATOR tool was used in addition. Weighted Euclidian distances (left) and Pearson correlation coefficients (right) were calculated between the different approaches for every sample, and the means of those values across all samples are represented as clustered heatmaps. (B) As in (A); the genome-wide copy number plots for sample T3 are displayed for the indicated analysis methods, with segmentation values depicted in red.
Figure 4
Copy number detection in the absence of a reference. (A) CopywriteR and onTarget methods were applied to WES data of melanoma PDX sample T99, either with or without C43 as a reference. Genome-wide copy number profiles are shown, with segmentation values (CBS) depicted in red. (B) CBS-derived segmentation values of the analysis in (A) are represented in a heatmap. (C) Segmentation values of all six melanoma PDX samples were treated as in (A) and (B), and the weighted Euclidian distances and Pearson correlation coefficients were calculated for every sample between the different methods. The means of those values across all samples are represented as clustered heatmaps.
Figure 5
CopywriteR is widely applicable. (A) Sample T97 (FFPE) was subjected to WES, and copy number profiles relative to C41 (fresh frozen reference material) are displayed for onTarget and CopywriteR methods, with segmentation values (CBS) depicted in red (left panel: whole-genome; right panel: chromosome 9). (B, left panel) ChIPseq data were obtained from ChIP experiments on the MCF7 cell line with the indicated set of antibodies, or from the relevant input control. Copy number data were extracted using CopywriteR, and further analyzed employing CBS. Segmentation values are represented as a heatmap. (B, right panel) Data were analyzed as for the left panel. ChIPseq data were obtained from ChIP experiments on ER+ breast cancer with ER-antibodies (E), or from the relevant input (I) control. (C, left panel) A set of matched pre- and post-vemurafenib treatment melanoma samples were subjected to targeted sequencing on a 1,977-gene panel. Copy number information was extracted using CopywriteR and example regions of the resulting copy number profiles are presented, with segmentation values (CBS) depicted in red. (C, right panel) Segmentation values were plotted as a heatmap for the pre/post-treatment pairs.
Similar articles
- Evaluation of somatic copy number estimation tools for whole-exome sequencing data.
Nam JY, Kim NK, Kim SC, Joung JG, Xi R, Lee S, Park PJ, Park WY. Nam JY, et al. Brief Bioinform. 2016 Mar;17(2):185-92. doi: 10.1093/bib/bbv055. Epub 2015 Jul 25. Brief Bioinform. 2016. PMID: 26210357 Free PMC article. - Determining multiallelic complex copy number and sequence variation from high coverage exome sequencing data.
Forni D, Martin D, Abujaber R, Sharp AJ, Sironi M, Hollox EJ. Forni D, et al. BMC Genomics. 2015 Nov 2;16:891. doi: 10.1186/s12864-015-2123-y. BMC Genomics. 2015. PMID: 26526070 Free PMC article. - SavvyCNV: Genome-wide CNV calling from off-target reads.
Laver TW, De Franco E, Johnson MB, Patel KA, Ellard S, Weedon MN, Flanagan SE, Wakeling MN. Laver TW, et al. PLoS Comput Biol. 2022 Mar 16;18(3):e1009940. doi: 10.1371/journal.pcbi.1009940. eCollection 2022 Mar. PLoS Comput Biol. 2022. PMID: 35294448 Free PMC article. - Exome sequence read depth methods for identifying copy number changes.
Kadalayil L, Rafiq S, Rose-Zerilli MJ, Pengelly RJ, Parker H, Oscier D, Strefford JC, Tapper WJ, Gibson J, Ennis S, Collins A. Kadalayil L, et al. Brief Bioinform. 2015 May;16(3):380-92. doi: 10.1093/bib/bbu027. Epub 2014 Aug 28. Brief Bioinform. 2015. PMID: 25169955 Review. - Free-access copy-number variant detection tools for targeted next-generation sequencing data.
Roca I, González-Castro L, Fernández H, Couce ML, Fernández-Marmiesse A. Roca I, et al. Mutat Res Rev Mutat Res. 2019 Jan-Mar;779:114-125. doi: 10.1016/j.mrrev.2019.02.005. Epub 2019 Feb 23. Mutat Res Rev Mutat Res. 2019. PMID: 31097148 Review.
Cited by
- Subtype-specific and co-occurring genetic alterations in B-cell non-Hodgkin lymphoma.
Ma MCJ, Tadros S, Bouska A, Heavican T, Yang H, Deng Q, Moore D, Akhter A, Hartert K, Jain N, Showell J, Ghosh S, Street L, Davidson M, Carey C, Tobin J, Perumal D, Vose JM, Lunning MA, Sohani AR, Chen BJ, Buckley S, Nastoupil LJ, Davis RE, Westin JR, Fowler NH, Parekh S, Gandhi M, Neelapu S, Stewart D, Bhalla K, Iqbal J, Greiner T, Rodig SJ, Mansoor A, Green MR. Ma MCJ, et al. Haematologica. 2022 Mar 1;107(3):690-701. doi: 10.3324/haematol.2020.274258. Haematologica. 2022. PMID: 33792219 Free PMC article. - The effect of missing data on evolutionary analysis of sequence capture bycatch, with application to an agricultural pest.
Featherstone LA, McGaughran A. Featherstone LA, et al. Mol Genet Genomics. 2024 Feb 21;299(1):11. doi: 10.1007/s00438-024-02097-7. Mol Genet Genomics. 2024. PMID: 38381254 Free PMC article. - Transcription Factor NFIB Is a Driver of Small Cell Lung Cancer Progression in Mice and Marks Metastatic Disease in Patients.
Semenova EA, Kwon MC, Monkhorst K, Song JY, Bhaskaran R, Krijgsman O, Kuilman T, Peters D, Buikhuisen WA, Smit EF, Pritchard C, Cozijnsen M, van der Vliet J, Zevenhoven J, Lambooij JP, Proost N, van Montfort E, Velds A, Huijbers IJ, Berns A. Semenova EA, et al. Cell Rep. 2016 Jul 19;16(3):631-43. doi: 10.1016/j.celrep.2016.06.020. Epub 2016 Jun 30. Cell Rep. 2016. PMID: 27373156 Free PMC article. - The TP53 mutation rate differs in breast cancers that arise in women with high or low mammographic density.
Cheasley D, Devereux L, Hughes S, Nickson C, Procopio P, Lee G, Li N, Pridmore V, Elder K, Bruce Mann G, Kader T, Rowley SM, Fox SB, Byrne D, Saunders H, Fujihara KM, Lim B, Gorringe KL, Campbell IG. Cheasley D, et al. NPJ Breast Cancer. 2020 Aug 7;6:34. doi: 10.1038/s41523-020-00176-7. eCollection 2020. NPJ Breast Cancer. 2020. PMID: 32802943 Free PMC article. - Targeting pyrimidine synthesis accentuates molecular therapy response in glioblastoma stem cells.
Wang X, Yang K, Wu Q, Kim LJY, Morton AR, Gimple RC, Prager BC, Shi Y, Zhou W, Bhargava S, Zhu Z, Jiang L, Tao W, Qiu Z, Zhao L, Zhang G, Li X, Agnihotri S, Mischel PS, Mack SC, Bao S, Rich JN. Wang X, et al. Sci Transl Med. 2019 Aug 7;11(504):eaau4972. doi: 10.1126/scitranslmed.aau4972. Sci Transl Med. 2019. PMID: 31391321 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases