Identifying multi-locus chromatin contacts in human cells using tethered multiple 3C - PubMed (original) (raw)
Identifying multi-locus chromatin contacts in human cells using tethered multiple 3C
Ferhat Ay et al. BMC Genomics. 2015.
Abstract
Background: Several recently developed experimental methods, each an extension of the chromatin conformation capture (3C) assay, have enabled the genome-wide profiling of chromatin contacts between pairs of genomic loci in 3D. Especially in complex eukaryotes, data generated by these methods, coupled with other genome-wide datasets, demonstrated that non-random chromatin folding correlates strongly with cellular processes such as gene expression and DNA replication.
Results: We describe a genome architecture assay, tethered multiple 3C (TM3C), that maps genome-wide chromatin contacts via a simple protocol of restriction enzyme digestion and religation of fragments upon agarose gel beads followed by paired-end sequencing. In addition to identifying contacts between pairs of loci, TM3C enables identification of contacts among more than two loci simultaneously. We use TM3C to assay the genome architectures of two human cell lines: KBM7, a near-haploid chronic leukemia cell line, and NHEK, a normal diploid human epidermal keratinocyte cell line. We confirm that the contact frequency maps produced by TM3C exhibit features characteristic of existing genome architecture datasets, including the expected scaling of contact probabilities with genomic distance, megabase scale chromosomal compartments and sub-megabase scale topological domains. We also confirm that TM3C captures several known cell type-specific contacts, ploidy shifts and translocations, such as Philadelphia chromosome formation (Ph+) in KBM7. We confirm a subset of the triple contacts involving the IGF2-H19 imprinting control region (ICR) using PCR analysis for KBM7 cells. Our genome-wide analysis of pairwise and triple contacts demonstrates their preference for linking open chromatin regions to each other and for linking regions with higher numbers of DNase hypersensitive sites (DHSs) to each other. For near-haploid KBM7 cells, we infer whole genome 3D models that exhibit clustering of small chromosomes with each other and large chromosomes with each other, consistent with previous studies of the genome architectures of other human cell lines.
Conclusion: TM3C is a simple protocol for ascertaining genome architecture and can be used to identify simultaneous contacts among three or four loci. Application of TM3C to a near-haploid human cell line revealed large-scale features of chromosomal organization and multi-way chromatin contacts that preferentially link regions of open chromatin.
Figures
Figure 1
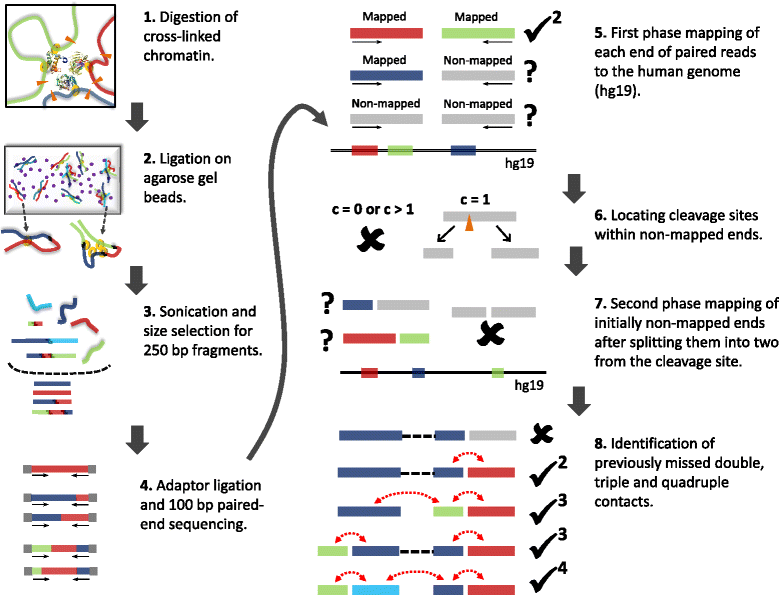
Overview of TM3C experimental protocol and mapping of paired-end reads to human genome. 1. Cells are treated with formaldehyde, covalently crosslinking proteins to one another and to the DNA. The DNA is then digested with either a single 4-cutter enzyme (DpnII) or a cocktail of enzymes (AluI, DpnII, MspI, and NlaIII). 2. Melted low-melting agarose solution is added to the digested nuclei to tether the DNA to agarose beads. Thin strings of the hot nuclei plus agarose solution is then transferred to an ice-cold ligation cocktail overnight. 3. After reversal of formaldehyde crosslinks and purification via gel extraction, the TM3C molecules are sonicated and size-selected for 250 bp fragments. 4. Size-selected fragments are paired-end sequenced (100 bp per end) after addition of sequencing adaptors. 5. Each end of paired-end reads are mapped to human reference genome. If both ends are mapped then the pair is considered a double and retained because it is informative for genome architecture. 6. Read ends that do not map to the reference genome are identified and segregated according to the number of cleavage sites they contain for the restriction enzyme(s) used for digestion. 7. Reads with exactly one cleavage site are considered for the second phase of mapping. These reads are split into two from the cleavage site and each of these two pieces are mapped back to the reference genome. 8. Read pairs with either one or both ends not mapped in the first mapping phase are reconsidered after second phase. Depending on how many pieces stemming from the original reads are mapped in the second phase, such pairs lead to either no informative contacts, doubles, triples or quadruples.
Figure 2
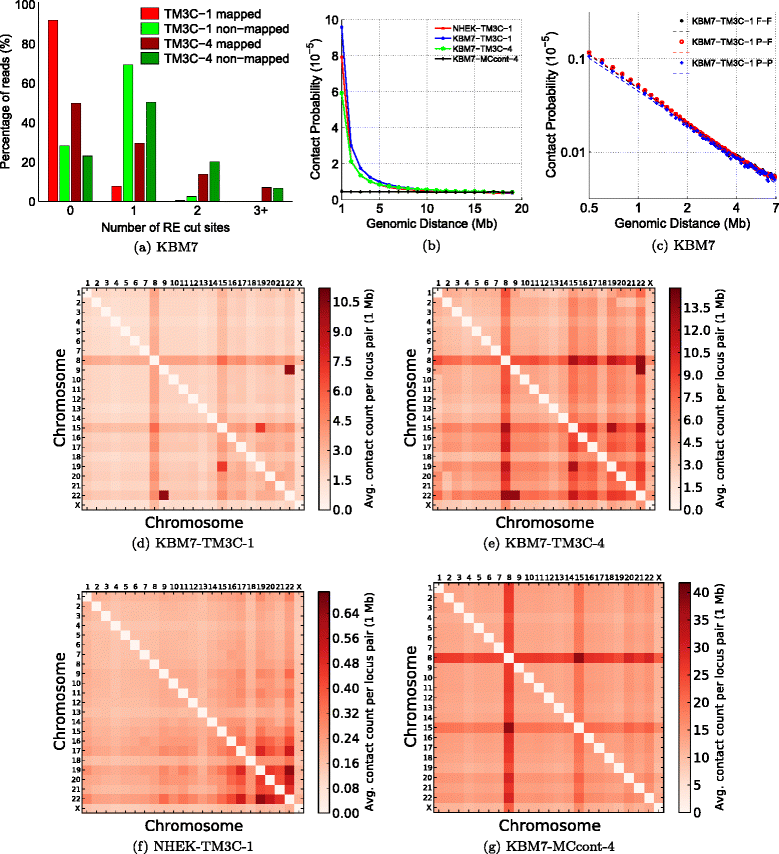
Consistency of TM3C data with known organizational principles and KBM7 karyotype. (a) Number of RE cut sites within reads that are fully mapped and nonmapped in the first phase mapping for KBM7 libraries. (b) Scaling of contact probability with genomic distance for three crosslinked libraries and one non-crosslinked control library. (c) Scaling of contact probability in log–log scale for three different sets of contacts identified in KBM7-TM3C-1 library. Pairwise chromosome contact matrices for (d) KBM7-TM3C-1, (e) KBM7-TM3C-4, (f) NHEK-TM3C-1 and (g) KBM7-MCcont-4 libraries. For these plots contact counts are averaged over all pairs of mappable 1 Mb windows between the two chromosomes.
Figure 3
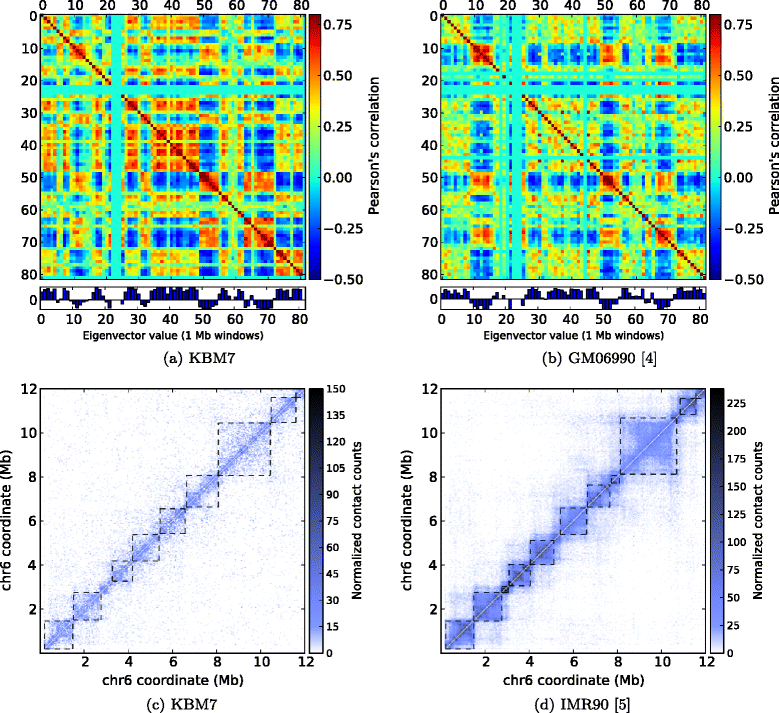
Comparison of TM3C data with existing genome architecture datasets. Eigenvalue decomposition to identify open/closed chromatin compartments of chromosome 17 (a) from the KBM7 cell line assayed by TM3C and (b) from GM06990 cell line assayed by Hi-C [4]. Topological domain calls and contact count heatmaps of a 6 Mb region of chromosome 6 (c) for the KBM7 cell line assayed by TM3C and (d) for the IMR90 cell line assayed by Hi-C [5].
Figure 4
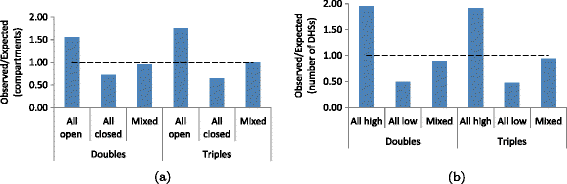
Genome-wide characterization of triple contacts. (a) Observed over expected percentages of double and triple contacts that link 1 Mb regions with the same (either open or closed) or different (mixed) compartment labels for the KBM7-TM3C-1 library (Methods). Both double and triple contacts prefer to link open compartments to each other with triples showing slightly more enrichment for this trend. (b) Similar percentages as in (a) but when 1 Mb windows are segregated according to the number of DHSs they contain (Methods). Contacts linking regions with higher numbers of DHSs than the median number are enriched within the doubles and the triples of the KBM7-TM3C-1 library. Due to lack of DNase data for KBM7 cells, we use data from six other human cell lines for this analysis. Since the results are very similar among different cell lines, here we only plot the results for K562 which is also a leukemia cell line.
Figure 5
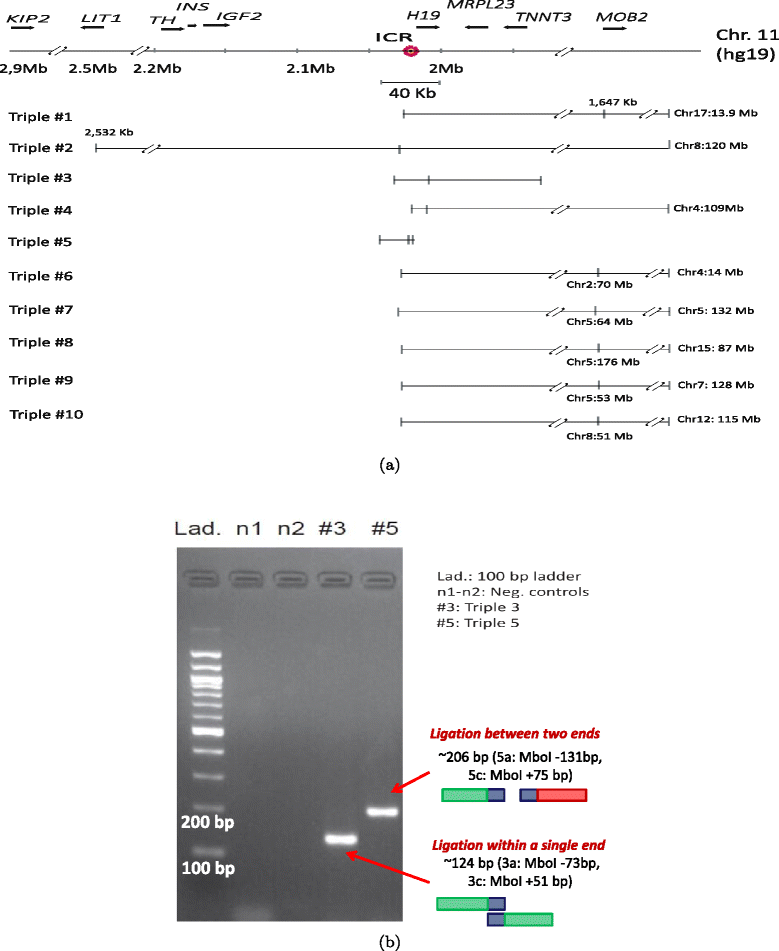
Validation of triples using PCR. (a) Ten triples extracted from the KBM7-TM3C-1 library that have at least one of their three ends in the 40 kb region surrounding the imprinting control region (ICR) of IGF2 and H19 genes. These triples involve short- and long-range contacts within chromosome 11 which are all indicated by tick marks with coordinates in kilobases (kb) displayed only for long-range contacts. Interchromosomal contacts with other chromosomes are indicated by the chromosome identifier followed by the coordinate in megabases (Mb). Orientation of the displayed locus is in the direction of IGF2 and H19 transcription. (b) PCR verification of pairwise contacts from triples 3 and 5. One pair of forward/reverse primers is used for each gel (Additional file 1: Table S1).
Figure 6
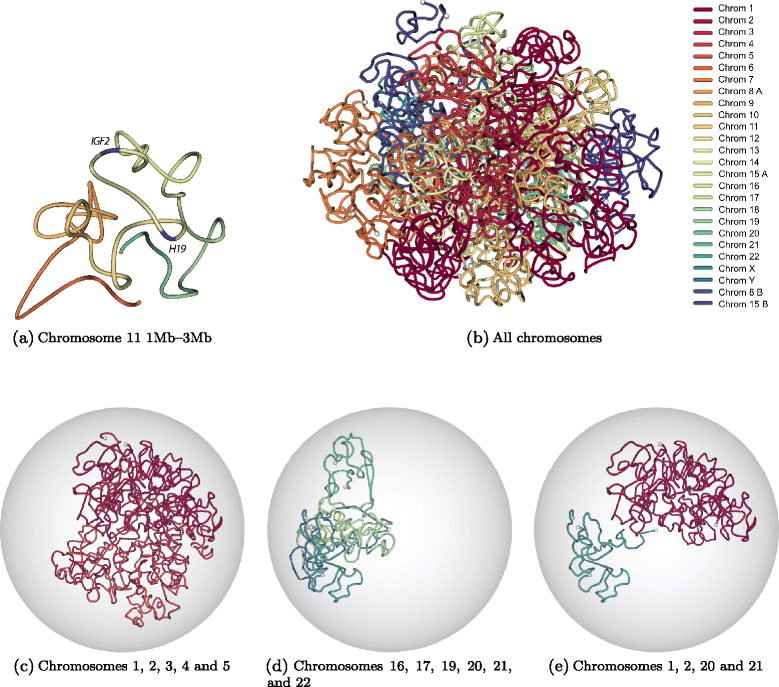
Three-dimensional modeling of KBM7 genome architecture. (a) Three-dimensional structure of the 2 Mb region of chromosome 11 (chr11:1,000,000-3,000,000) which is centered around IGF2-H19 imprinting control region. This structure is inferred from normalized contact counts of KBM7-TM3C-1 data at 40 kb resolution using the Poisson model from Varoquaux et al. [27]. (b) Three-dimensional structure of the KBM7 genome, which is haploid for all chromosomes other than diploid chromosome 8 (8A, 8B) and partially diploid chromosome 15 (15A, 15B) (see Methods for details of the 3D inference). Different colors represent different chromosomes, and white balls represent chromosome ends. Same 3D structure as (b) when confined to (c) only a subset of long chromosomes, (d) only a subset of small chromosomes, (e) two small and two large chromosomes.
Similar articles
- Hi-C: a method to study the three-dimensional architecture of genomes.
van Berkum NL, Lieberman-Aiden E, Williams L, Imakaev M, Gnirke A, Mirny LA, Dekker J, Lander ES. van Berkum NL, et al. J Vis Exp. 2010 May 6;(39):1869. doi: 10.3791/1869. J Vis Exp. 2010. PMID: 20461051 Free PMC article. - Tethered Chromosome Conformation Capture Sequencing in Triticeae: A Valuable Tool for Genome Assembly.
Himmelbach A, Walde I, Mascher M, Stein N. Himmelbach A, et al. Bio Protoc. 2018 Aug 5;8(15):e2955. doi: 10.21769/BioProtoc.2955. eCollection 2018 Aug 5. Bio Protoc. 2018. PMID: 34395764 Free PMC article. - Hi-C 3.0: Improved Protocol for Genome-Wide Chromosome Conformation Capture.
Lafontaine DL, Yang L, Dekker J, Gibcus JH. Lafontaine DL, et al. Curr Protoc. 2021 Jul;1(7):e198. doi: 10.1002/cpz1.198. Curr Protoc. 2021. PMID: 34286910 Free PMC article. - The macro and micro of chromosome conformation capture.
Goel VY, Hansen AS. Goel VY, et al. Wiley Interdiscip Rev Dev Biol. 2021 Nov;10(6):e395. doi: 10.1002/wdev.395. Epub 2020 Sep 28. Wiley Interdiscip Rev Dev Biol. 2021. PMID: 32987449 Free PMC article. Review. - Chromatin structural elements and chromosomal translocations in leukemia.
Zhang Y, Rowley JD. Zhang Y, et al. DNA Repair (Amst). 2006 Sep 8;5(9-10):1282-97. doi: 10.1016/j.dnarep.2006.05.020. Epub 2006 Aug 7. DNA Repair (Amst). 2006. PMID: 16893685 Review.
Cited by
- 4D nucleomes in single cells: what can computational modeling reveal about spatial chromatin conformation?
Sekelja M, Paulsen J, Collas P. Sekelja M, et al. Genome Biol. 2016 Apr 7;17:54. doi: 10.1186/s13059-016-0923-2. Genome Biol. 2016. PMID: 27052789 Free PMC article. Review. - The role of 3D genome organization in disease: From compartments to single nucleotides.
Chakraborty A, Ay F. Chakraborty A, et al. Semin Cell Dev Biol. 2019 Jun;90:104-113. doi: 10.1016/j.semcdb.2018.07.005. Epub 2018 Jul 17. Semin Cell Dev Biol. 2019. PMID: 30017907 Free PMC article. Review. - Large scale genomic reorganization of topological domains at the HoxD locus.
Fabre PJ, Leleu M, Mormann BH, Lopez-Delisle L, Noordermeer D, Beccari L, Duboule D. Fabre PJ, et al. Genome Biol. 2017 Aug 7;18(1):149. doi: 10.1186/s13059-017-1278-z. Genome Biol. 2017. PMID: 28784160 Free PMC article. - The Nodewalk assay to quantitate chromatin fiber interactomes in very small cell populations.
Vestlund J, Sumida N, Mehmood R, Bhartiya D, Wu S, Göndör A. Vestlund J, et al. Nat Protoc. 2023 Mar;18(3):755-782. doi: 10.1038/s41596-022-00774-8. Epub 2022 Nov 25. Nat Protoc. 2023. PMID: 36434098 Review. - Extensive Heterogeneity and Intrinsic Variation in Spatial Genome Organization.
Finn EH, Pegoraro G, Brandão HB, Valton AL, Oomen ME, Dekker J, Mirny L, Misteli T. Finn EH, et al. Cell. 2019 Mar 7;176(6):1502-1515.e10. doi: 10.1016/j.cell.2019.01.020. Epub 2019 Feb 21. Cell. 2019. PMID: 30799036 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous