Complete genomes reveal signatures of demographic and genetic declines in the woolly mammoth - PubMed (original) (raw)
. 2015 May 18;25(10):1395-400.
doi: 10.1016/j.cub.2015.04.007. Epub 2015 Apr 23.
Swapan Mallick 2, Pontus Skoglund 3, Jacob Enk 4, Nadin Rohland 5, Heng Li 5, Ayça Omrak 6, Sergey Vartanyan 7, Hendrik Poinar 8, Anders Götherström 6, David Reich 2, Love Dalén 9
Affiliations
- PMID: 25913407
- PMCID: PMC4439331
- DOI: 10.1016/j.cub.2015.04.007
Complete genomes reveal signatures of demographic and genetic declines in the woolly mammoth
Eleftheria Palkopoulou et al. Curr Biol. 2015.
Abstract
The processes leading up to species extinctions are typically characterized by prolonged declines in population size and geographic distribution, followed by a phase in which populations are very small and may be subject to intrinsic threats, including loss of genetic diversity and inbreeding. However, whether such genetic factors have had an impact on species prior to their extinction is unclear; examining this would require a detailed reconstruction of a species' demographic history as well as changes in genome-wide diversity leading up to its extinction. Here, we present high-quality complete genome sequences from two woolly mammoths (Mammuthus primigenius). The first mammoth was sequenced at 17.1-fold coverage and dates to ∼4,300 years before present, representing one of the last surviving individuals on Wrangel Island. The second mammoth, sequenced at 11.2-fold coverage, was obtained from an ∼44,800-year-old specimen from the Late Pleistocene population in northeastern Siberia. The demographic trajectories inferred from the two genomes are qualitatively similar and reveal a population bottleneck during the Middle or Early Pleistocene, and a more recent severe decline in the ancestors of the Wrangel mammoth at the end of the last glaciation. A comparison of the two genomes shows that the Wrangel mammoth has a 20% reduction in heterozygosity as well as a 28-fold increase in the fraction of the genome that comprises runs of homozygosity. We conclude that the population on Wrangel Island, which was the last surviving woolly mammoth population, was subject to reduced genetic diversity shortly before it became extinct.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures
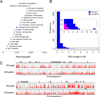
Figure 1. Geographic location and dating of samples, mapping statistics of the two genomic libraries and inference of population size changes through time
(A) Map indicating the sites from where the mammoth samples were collected. The red dashed line indicates the approximate extent of the coastline during the Last Glacial Maximum. (B) Sample dating information and mapping statistics of the two libraries. 14C date ± error refers to the radiocarbon age of each specimen and associated standard error. Median calibrated date refers to the median estimate of the calibrated radiocarbon date. Average sequence length (bp) refers to aligned sequences only. (C) Population size history inferred using the PSMC method. Time is given in units of divergence per base pair on the lower x-axis and years on the upper x-axis assuming the substitution rate estimated in this study based on the age difference between the two samples (the range given in parentheses takes into account the uncertainty of the rate estimate as well the range of rate estimates obtained from paleontological calibration; see Table 1). The PSMC curves of the Oimyakon genome and the pseudo-diploid chromosome X are empirically corrected for missing heterozygotes (false negatives [FN] = 30%) and are shifted along the x-axis so that the former is aligned to the curve of the Wrangel genome and the latter ends at ~24,500 years ago, the average age of the two individuals (which was converted in units of divergence based on the mean substitution rate estimated in this study). The Ne of the PSMC curve of the pseudo-diploid chromosome X was scaled by 0.75. The Eemian interglacial period and the Pleistocene/Holocene transition are indicated by grey vertical bars assuming the mean substitution rate estimated in this study. See also Table S1 and Figure S1.
Figure 2. Population split time estimate between the Wrangel and Oimyakon populations
The probability F(A|B) of observing a derived allele in population A (Wrangel) at a heterozygous site in population B (Oimyakon) is obtained by simulating the history of population B (Oimyakon) as inferred using the PSMC method. Polarization of the alleles was based on the African savanna elephant that was assumed to carry the ancestral state. The vertical dotted lines indicate the split times between the Wrangel and Oimyakon populations that encompass the confidence interval (CI) of the observed F(Wrangel|Oimyakon). Time on the x-axis represents the population split time before the death of the Oimyakon individual and is scaled by Ne and generation time, where Ne indicates the terminal effective population size of the Oimyakon individual as inferred using the PSMC analysis. See also Figure S2.
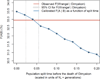
Figure 3. Estimates of genome-wide heterozygosity and runs of homozygosity
(A) Comparison of genome-wide heterozygosity estimates in different taxa/populations [–31] and the two woolly mammoths. See also Table S2. (B) Histogram of the lengths (in Mb) of ROHs that are longer than 0.5Mb within the Wrangel (blue) and Oimyakon (purple) genomes. The inset figure is a close-up of figure 3B with a different scale of the y-axis. (C) Inferred TMRCA for the two alleles of a single individual along 106Mb of chromosome 1 and chromosome 3, with Wrangel on top and Oimyakon at the bottom. The y-axis shows the TMRCA and the x-axis positions along the chromosome. Grey bars indicate regions longer than 1Mb that are inferred to have coalesced recently by the PSMC method and represent ROHs. See also Figure S4.
Similar articles
- Excess of genomic defects in a woolly mammoth on Wrangel island.
Rogers RL, Slatkin M. Rogers RL, et al. PLoS Genet. 2017 Mar 2;13(3):e1006601. doi: 10.1371/journal.pgen.1006601. eCollection 2017 Mar. PLoS Genet. 2017. PMID: 28253255 Free PMC article. - Temporal dynamics of woolly mammoth genome erosion prior to extinction.
Dehasque M, Morales HE, Díez-Del-Molino D, Pečnerová P, Chacón-Duque JC, Kanellidou F, Muller H, Plotnikov V, Protopopov A, Tikhonov A, Nikolskiy P, Danilov GK, Giannì M, van der Sluis L, Higham T, Heintzman PD, Oskolkov N, Gilbert MTP, Götherström A, van der Valk T, Vartanyan S, Dalén L. Dehasque M, et al. Cell. 2024 Jul 11;187(14):3531-3540.e13. doi: 10.1016/j.cell.2024.05.033. Epub 2024 Jun 27. Cell. 2024. PMID: 38942016 - The flickering genes of the last mammoths.
Thomas MG. Thomas MG. Mol Ecol. 2012 Jul;21(14):3379-81. doi: 10.1111/j.1365-294x.2012.05594.x. Mol Ecol. 2012. PMID: 22953331 - Cloning the mammoth: a complicated task or just a dream?
Loi P, Saragusty J, Ptak G. Loi P, et al. Adv Exp Med Biol. 2014;753:489-502. doi: 10.1007/978-1-4939-0820-2_19. Adv Exp Med Biol. 2014. PMID: 25091921 Review. - Resurrecting ancient animal genomes: the extinct moa and more.
Huynen L, Millar CD, Lambert DM. Huynen L, et al. Bioessays. 2012 Aug;34(8):661-9. doi: 10.1002/bies.201200040. Epub 2012 Jun 6. Bioessays. 2012. PMID: 22674514 Review.
Cited by
- Detectability of runs of homozygosity is influenced by analysis parameters and population-specific demographic history.
Silva GAA, Harder AM, Kirksey KB, Mathur S, Willoughby JR. Silva GAA, et al. PLoS Comput Biol. 2024 Oct 31;20(10):e1012566. doi: 10.1371/journal.pcbi.1012566. eCollection 2024 Oct. PLoS Comput Biol. 2024. PMID: 39480880 Free PMC article. - Heterochronous mitogenomes shed light on the Holocene history of the Scandinavian brown bear.
Feinauer IS, Lord E, von Seth J, Xenikoudakis G, Ersmark E, Dalén L, Meleg IN. Feinauer IS, et al. Sci Rep. 2024 Oct 22;14(1):24917. doi: 10.1038/s41598-024-75028-6. Sci Rep. 2024. PMID: 39438503 Free PMC article. - Whole genomes from the extinct Xerces Blue butterfly can help identify declining insect species.
de-Dios T, Fontsere C, Renom P, Stiller J, Llovera L, Uliano-Silva M, Sánchez-Gracia A, Wright C, Lizano E, Caballero B, Navarro A, Civit S, Robbins RK, Blaxter M, Marquès T, Vila R, Lalueza-Fox C. de-Dios T, et al. Elife. 2024 Oct 4;12:RP87928. doi: 10.7554/eLife.87928. Elife. 2024. PMID: 39365295 Free PMC article. - Demography and environment modulate the effects of genetic diversity on extinction risk in a butterfly metapopulation.
DiLeo MF, Nair A, Kardos M, Husby A, Saastamoinen M. DiLeo MF, et al. Proc Natl Acad Sci U S A. 2024 Aug 13;121(33):e2309455121. doi: 10.1073/pnas.2309455121. Epub 2024 Aug 8. Proc Natl Acad Sci U S A. 2024. PMID: 39116125 Free PMC article. - Sirenian genomes illuminate the evolution of fully aquatic species within the mammalian superorder afrotheria.
Tian R, Zhang Y, Kang H, Zhang F, Jin Z, Wang J, Zhang P, Zhou X, Lanyon JM, Sneath HL, Woolford L, Fan G, Li S, Seim I. Tian R, et al. Nat Commun. 2024 Jul 2;15(1):5568. doi: 10.1038/s41467-024-49769-x. Nat Commun. 2024. PMID: 38956050 Free PMC article.
References
- Caughley G. Directions in Conservation Biology. J. Anim. Ecol. 1994;63:215–244.
- Jamieson IG, Allendorf FW. How does the 50/500 rule apply to MVPs? Trends in Ecology & Evolution. 2012;27:578–584. - PubMed
- Stuart AJ, Kosintsev PA, Higham TFG, Lister AM. Pleistocene to Holocene extinction dynamics in giant deer and woolly mammoth. Nature. 2004;431:684–689. - PubMed
- Vartanyan SL, Garutt VE, Sher AV. Holocene dwarf mammoths from Wrangel Island in the Siberian Arctic. Nature. 1993;362:337–340. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources