The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3 - PubMed (original) (raw)
. 2015 May 14;521(7551):232-6.
doi: 10.1038/nature14443. Epub 2015 Apr 27.
Chun-Kan Chen 1, Amy Chow 1, Christine F Surka 1, Christina Tran 1, Patrick McDonel 2, Amy Pandya-Jones 3, Mario Blanco 1, Christina Burghard 1, Annie Moradian 4, Michael J Sweredoski 4, Alexander A Shishkin 1, Julia Su 1, Eric S Lander 2, Sonja Hess 4, Kathrin Plath 3, Mitchell Guttman 1
Affiliations
- PMID: 25915022
- PMCID: PMC4516396
- DOI: 10.1038/nature14443
The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3
Colleen A McHugh et al. Nature. 2015.
Abstract
Many long non-coding RNAs (lncRNAs) affect gene expression, but the mechanisms by which they act are still largely unknown. One of the best-studied lncRNAs is Xist, which is required for transcriptional silencing of one X chromosome during development in female mammals. Despite extensive efforts to define the mechanism of Xist-mediated transcriptional silencing, we still do not know any proteins required for this role. The main challenge is that there are currently no methods to comprehensively define the proteins that directly interact with a lncRNA in the cell. Here we develop a method to purify a lncRNA from cells and identify proteins interacting with it directly using quantitative mass spectrometry. We identify ten proteins that specifically associate with Xist, three of these proteins--SHARP, SAF-A and LBR--are required for Xist-mediated transcriptional silencing. We show that SHARP, which interacts with the SMRT co-repressor that activates HDAC3, is not only essential for silencing, but is also required for the exclusion of RNA polymerase II (Pol II) from the inactive X. Both SMRT and HDAC3 are also required for silencing and Pol II exclusion. In addition to silencing transcription, SHARP and HDAC3 are required for Xist-mediated recruitment of the polycomb repressive complex 2 (PRC2) across the X chromosome. Our results suggest that Xist silences transcription by directly interacting with SHARP, recruiting SMRT, activating HDAC3, and deacetylating histones to exclude Pol II across the X chromosome.
Figures
Extended Data Figure 1. RAP-MS recovers and enriches the majority of Xist RNA from mouse ES cells, and these cells can be efficiently labeled with SILAC
(a) RT-qPCR measuring the percentage of the total cellular Xist or 18S recovered after RAP-MS of Xist. Values are computed as the amount of each RNA in the elution divided by the amount of RNA in the starting (“input”) lysate material. Error bars represent the standard error of the mean from 5 biological replicates. (b) Enrichment of Xist after RAP-MS captures from pSM33 cells as measured by qPCR. Bars indicate RNA levels of Xist, 18S, and Oct4 after purification of Xist, normalized to RNA in input sample. Each bar represents the RNA levels of Xist, 18S, and Oct4 after purification of Xist, normalized to RNA in input sample, from 3 biological replicates. (c) SILAC labeling efficiency of a representative culture of pSM33 mouse ES cells after 10 days of growth (3 cell passages) in SILAC medium. Peptides were analyzed by mass spectrometry, and values indicate the fraction of identified peptides with heavy-label incorporation with different levels of peptide labeling (shown in bins).
Extended Data Figure 2. RAP-MS identifies proteins that are known to directly interact with specific ncRNAs, and separates specific RNA interacting proteins from background proteins
(a) SILAC ratios of top proteins enriched in the RAP-MS U1 snRNA, 18S rRNA, and 45S pre-rRNA experiments. (b) SILAC ratio plot of replicate captures of U1 snRNA versus 18S rRNA from one of two biologically independent label-swap experiments. Proteins associated with U1 are consistently found in U1 samples, both light and heavy labeled (top right quadrant), and proteins specifically associated with 18S are consistently identified in 18S, both light and heavy (lower left quadrant). Background contaminant proteins have low enrichments (center of panel) or are consistently found in the light channel and do not replicate between experiments (i.e. keratin, streptavidin). (c) SILAC ratio plot of replicate captures of U1 snRNA versus 45S pre-rRNA from one label-swap experiment. Proteins that are known to associate with 45S pre-rRNA are consistently identified in 45S captures.
Extended Data Figure 3. Immunoprecipitation of the identified Xist-interacting proteins confirms Xist RNA interaction
RNA immunoprecipitation experiments were performed for seven Xist-interacting proteins (black bars), two control RNA binding proteins that were not identified by RAP-MS and IgG (gray bars) in UV-crosslinked cell lysate after 6 hours of Xist induction by doxycycline addition (Methods). The RNA associated with each protein was measured and enrichment levels were computed relative to the level of the RNA in total cellular input and normalized to the total efficiency of capture in each sample to allow for direct comparison across all IP experiments (Methods). (a) Enrichment of the Xist lncRNA after immunoprecipitation from a sample of pSM33 male cells (b) Immunoprecipitation of SHARP was performed from a sample of UV-crosslinked females ES cells that were treated with retinoic acid for 24 hours. The levels of recovered Xist lncRNA (black bars), Neat1 lncRNA (white bars), and 45S pre-ribosomal RNA (gray bars) were measured by RT-qPCR. Enrichment of each RNA after capture with anti-SHARP antibody was calculated relative to the level of RNA captured with IgG control antibody. (c) The enrichment of various lncRNAs after immunoprecipitation in pSM33 male cells – including Neat1, Malat1, Firre, and Tug1 – are shown. (d) The enrichment of various mRNA controls after immunoprecipitation in pSM33 male cells – including Oct4, Nanog, Stat3, and Suz12 – are shown.
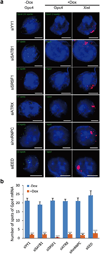
Extended Data Figure 4. Previously identified proteins associated with XCI are not required for Xist-mediated transcriptional silencing
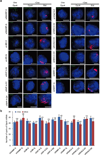
(a) To confirm the specificity of our assay, we tested the function of several proteins that were previously identified to associate with Xist, but not to silence transcription, for their role in transcriptional silencing in our inducible male ES cells prior to Xist induction (−Dox; left) or after Xist induction for 16 hours (+Dox; middle and right). Representative images are shown after knockdown of each protein – DAPI (blue), Xist (red), and Gpc4 (green). (b) Quantification of the copy number of Gpc4 before and after Xist induction upon treatment with different siRNAs. Error bars represent the standard error of the mean across 50 individual cells from one experiment. **** represents values with a p-value<0.001 between +Dox and −Dox cells based on an unpaired two-sample t-test. Scale bars on the images represent 5 µm. Importantly, while these proteins do not have a role in the initiation of transcriptional silencing, we do not mean to imply that they do not have other roles in XCI.
Extended Data Figure 5. SHARP, LBR, SAF-A, SMRT, and HDAC3 are required for Xist-mediated transcriptional silencing
(a) Representative images showing staining of DAPI (blue), Xist (red), and Gpc4 (green) for different siRNA knockdown in male ES cells prior to Xist induction (−Dox; left) or after Xist induction for 16 hours (+Dox; middle and right). (b) Quantification of the copy number of Gpc4 in −Dox and +Dox cells after knockdown with siRNAs targeting different mRNAs. Error bars represent the standard error of the mean across 50 individual cells from one experiment. NS: not significantly different between +Dox and −Dox cells; **** represents values with a p-value<0.001 between +Dox and –Dox cells based on an unpaired two-sample t-test. Scale bars on the images represent 5 µm. (c) Knockdown of SHARP, LBR, or SAF-A abrogates Xist-mediated gene silencing without causing pluripotency defects. Representative images showing staining of Nanog (cyan), Xist (red), and Gpc4 (green) upon knockdown of SHARP, LBR or SAF-A after 16 hours of Xist induction with doxycycline. Scale bars on the images represent 5 µm.
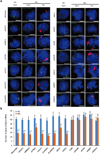
Extended Data Figure 6. SHARP is required for silencing many genes across the X-chromosome
(a) A diagram showing the locations of Xist (red), X-linked silenced genes (black), and X-linked escaped genes (green) along the X-chromosome. (b) Representative images showing staining of DAPI (blue), Xist (red), X-linked silenced genes (green), and X-linked escaped genes (yellow) upon knockdown of SHARP or control male ES cells prior to Xist induction (−Dox) or after Xist induction for 16 hours (+Dox). Knock of SHARP abolishes the silencing of Atrx, Gpc4, Rbmx, Smc1a and Mecp2, which are normally silenced upon Xist expression, but has no effect on Mid1 and Pir, which normally escape Xist-mediated silencing. The bar graphs show the quantification of the copy number of the mRNA for each gene for −Dox and +Dox cells upon transfection with SHARP siRNA or control siRNA; Error bars represent the standard error of the mean across 50 individual cells from one experiment, NS: not significantly different, **** represents values with a p-value<0.001, and ** represents values with a p-value<0.01 between +Dox and −Dox cells based on an unpaired two-sample t-test. Scale bars on the images represent 5 µm.
Extended Data Figure 7. Multiple independent siRNAs targeting SHARP, LBR, SAF-A, HDAC3, or SMRT demonstrate the same silencing defect
(a) Representative images showing staining of DAPI (blue), Xist (red), and Gpc4 (green) after knockdown of proteins using independent, non-overlapping, siRNA pools, or individual siRNA deconvoluted from the pool prior to Xist induction (−Dox; left) or after Xist induction for 16 hours (+Dox; middle and right). Cells were either transfected with the siRNA pool from Dharmacon (siRNA-D), Qiagen (siRNA-Q) or Ambion/Life Technologies (siRNA-A), or each individual siRNA deconvoluted from the pool from Dharmacon (siRNA-D1, 2, 3, 4) or Qiagen (siRNA-Q1, 2, 3, 4). Error bars represent the standard error of the mean across 50 individual cells from one experiment. NS: not significantly different between +Dox and −Dox cells based on an unpaired two-sample t-test. Scale bars on the images represent 5 µm. We excluded all siRNAs that did not reduce the targeted mRNA level by >70% (Methods). The sequences of deconvoluted siRNAs are shown in Supplementary Table 2.
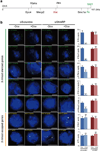
Extended Data Figure 8. SHARP, LBR, SAF-A, SMRT, and HDAC3 are required for transcriptional silencing in differentiating female ES cells
(a) Representative images showing staining of DAPI (blue), Xist (red), and Gpc4 (green) upon knockdown of specific proteins using different siRNAs in female ES cells prior to differentiation (−RA; left) or after differentiation for 24 hours (+RA; middle and right). (b) Quantification of the copy number of Gpc4 for −RA and +RA cells upon transfection with different siRNAs. Error bars represent the standard error across 50 individual cells from one experiment. NS: not significantly different between +RA and −RA cells; **** represents values with a p-value<0.001, ** represents values with a p-value<0.01, and * represents values with a p-value<0.05 between +RA and −RA cells based on an unpaired two-sample t-test. Scale bars on the images represent 5 µm.
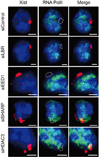
Extended Data Figure 9. SHARP is required for exclusion of RNA Polymerase II from the Xist-coated territory in differentiating female ES cells
Images of individual cells that are labeled with Xist (red), RNA Polymerase II (green), and DAPI (blue) across different siRNA conditions (rows) in female ES cells after 24 hours of retinoic acid treatment. The dashed white region represents the outlined Xist coated territory.
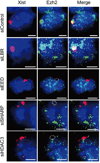
Extended Data Figure 10. SHARP is required for PRC2 recruitment across the Xist coated territory in differentiating female ES cells
Images of individual cells that are labeled with Xist (red), Ezh2 (green) and DAPI (blue) across different siRNA conditions (rows) in female ES cells after 24 hours of differentiation. The dashed white region represents the outlined Xist coated territory.
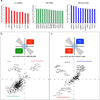
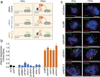
Figure 1. RAP-MS identifies direct Xist-interacting proteins
(a) A schematic overview of the RAP-MS method. (b) The SILAC ratio (Xist/U1) for each Xist-enriched protein identified by RAP-MS for one representative sample of four biological replicates. For SHARP and RBM15, the enrichment values are indicated above their bars. (c) Each Xist-interacting protein is shown (scaled to protein length). The locations of functional domains are shown.
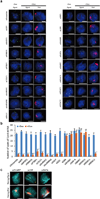
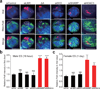
Figure 2. SHARP, LBR, and SAF-A are required for Xist-mediated gene silencing
(a) Screen for Xist-mediated gene silencing for knockdown of control (top), non-silencing proteins (middle), or silencing proteins (bottom). (b) Gpc4 mRNA levels after induction of Xist (+dox) normalized to Gpc4 levels before Xist induction (−dox). Error bars: standard error of the mean across 50 cells from one experiment. siCtrl: scrambled siRNA control. (c) Images of individual cells for two X-linked mRNAs, Gpc4 (green) and Atrx (red), and DAPI (blue) after treatment with different siRNAs (rows). The number of identified mRNAs is shown. Scale bars, 5 micrometers.
Figure 3. SHARP is required for exclusion of PolII from the Xist-coated territory
(a) Xist (red), PolII (green), and DAPI (blue) across different siRNA conditions (rows). Quantification of fluorescence intensity of PolII within Xist territory normalized to control siRNA levels for (b) male ES cells after 16 hours of doxycycline treatment and (c) female ES cells after 1 day of retinoic acid induced differentiation. Error bars: standard error of the mean across 50 cells from one experiment. NS: not significant, **** _p_-value<0.001 relative to siControl by unpaired two-sample t-test. ΔA: Genetic deletion of A-repeat. Scale bars, 5 micrometers.
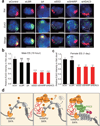
Figure 4. SHARP is required for PRC2 recruitment across the Xist-coated territory
(a) Xist (red), Ezh2 (green) and DAPI (blue) across siRNA conditions (rows). Quantification of Ezh2 levels within the defined Xist territory normalized to the levels in the control siRNA sample for (b) male ES cells and (c) differentiating female ES cells. Error bars: standard error of the mean across 50 cells from one experiment. NS: not significant, ***, _p_-value<0.005, **** _p_-value<0.001 relative to siControl by an unpaired two-sample t-test. Scale bars, 5 micrometers. (d) A model for Xist-mediated transcriptional silencing and recruitment of PRC2 across the X-chromosome.
Comment in
- Non-coding RNA: X-chromosome inactivation unravelled.
Zlotorynski E. Zlotorynski E. Nat Rev Mol Cell Biol. 2015 Jun;16(6):325. doi: 10.1038/nrm3998. Epub 2015 May 8. Nat Rev Mol Cell Biol. 2015. PMID: 25953617 No abstract available. - Molecular biology: Rap and chirp about X inactivation.
Roth A, Diederichs S. Roth A, et al. Nature. 2015 May 14;521(7551):170-1. doi: 10.1038/521170a. Nature. 2015. PMID: 25971508 No abstract available. - Non-coding RNA: X chromosome inactivation unravelled.
Zlotorynski E. Zlotorynski E. Nat Rev Genet. 2015 Jun;16(6):315. doi: 10.1038/nrg3955. Nat Rev Genet. 2015. PMID: 25982167 No abstract available.
Similar articles
- Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing.
Chen CK, Blanco M, Jackson C, Aznauryan E, Ollikainen N, Surka C, Chow A, Cerase A, McDonel P, Guttman M. Chen CK, et al. Science. 2016 Oct 28;354(6311):468-472. doi: 10.1126/science.aae0047. Epub 2016 Aug 4. Science. 2016. PMID: 27492478 - Xist reduction in breast cancer upregulates AKT phosphorylation via HDAC3-mediated repression of PHLPP1 expression.
Huang YS, Chang CC, Lee SS, Jou YS, Shih HM. Huang YS, et al. Oncotarget. 2016 Jul 12;7(28):43256-43266. doi: 10.18632/oncotarget.9673. Oncotarget. 2016. PMID: 27248326 Free PMC article. - hnRNPK Recruits PCGF3/5-PRC1 to the Xist RNA B-Repeat to Establish Polycomb-Mediated Chromosomal Silencing.
Pintacuda G, Wei G, Roustan C, Kirmizitas BA, Solcan N, Cerase A, Castello A, Mohammed S, Moindrot B, Nesterova TB, Brockdorff N. Pintacuda G, et al. Mol Cell. 2017 Dec 7;68(5):955-969.e10. doi: 10.1016/j.molcel.2017.11.013. Mol Cell. 2017. PMID: 29220657 Free PMC article. - Progress toward understanding chromosome silencing by Xist RNA.
Brockdorff N, Bowness JS, Wei G. Brockdorff N, et al. Genes Dev. 2020 Jun 1;34(11-12):733-744. doi: 10.1101/gad.337196.120. Genes Dev. 2020. PMID: 32482714 Free PMC article. Review. - The making of a Barr body: the mosaic of factors that eXIST on the mammalian inactive X chromosome.
Dixon-McDougall T, Brown C. Dixon-McDougall T, et al. Biochem Cell Biol. 2016 Feb;94(1):56-70. doi: 10.1139/bcb-2015-0016. Epub 2015 Jun 24. Biochem Cell Biol. 2016. PMID: 26283003 Review.
Cited by
- Synthetic tetracycline-controllable shRNA targeting long non-coding RNA HOXD-AS1 inhibits the progression of bladder cancer.
Li J, Zhuang C, Liu Y, Chen M, Chen Y, Chen Z, He A, Lin J, Zhan Y, Liu L, Xu W, Zhao G, Guo Y, Wu H, Cai Z, Huang W. Li J, et al. J Exp Clin Cancer Res. 2016 Jun 21;35(1):99. doi: 10.1186/s13046-016-0372-5. J Exp Clin Cancer Res. 2016. PMID: 27328915 Free PMC article. - X Inactivation Lessons from Differentiating Mouse Embryonic Stem Cells.
Pintacuda G, Cerase A. Pintacuda G, et al. Stem Cell Rev Rep. 2015 Oct;11(5):699-705. doi: 10.1007/s12015-015-9597-5. Stem Cell Rev Rep. 2015. PMID: 26198263 Free PMC article. Review. - RNA-protein interaction mapping via MS2- or Cas13-based APEX targeting.
Han S, Zhao BS, Myers SA, Carr SA, He C, Ting AY. Han S, et al. Proc Natl Acad Sci U S A. 2020 Sep 8;117(36):22068-22079. doi: 10.1073/pnas.2006617117. Epub 2020 Aug 24. Proc Natl Acad Sci U S A. 2020. PMID: 32839320 Free PMC article. - New insights into the functional role of retrotransposon dynamics in mammalian somatic cells.
Mangiavacchi A, Liu P, Della Valle F, Orlando V. Mangiavacchi A, et al. Cell Mol Life Sci. 2021 Jul;78(13):5245-5256. doi: 10.1007/s00018-021-03851-5. Epub 2021 May 14. Cell Mol Life Sci. 2021. PMID: 33990851 Free PMC article. Review. - SHIFTR enables the unbiased identification of proteins bound to specific RNA regions in live cells.
Aydin J, Gabel A, Zielinski S, Ganskih S, Schmidt N, Hartigan CR, Schenone M, Carr SA, Munschauer M. Aydin J, et al. Nucleic Acids Res. 2024 Mar 21;52(5):e26. doi: 10.1093/nar/gkae038. Nucleic Acids Res. 2024. PMID: 38281241 Free PMC article.
References
- Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat Rev Genet. 2011;12:542–553. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM007616/GM/NIGMS NIH HHS/United States
- T32GM07616/GM/NIGMS NIH HHS/United States
- DP5OD012190/OD/NIH HHS/United States
- DP2 OD001686/OD/NIH HHS/United States
- R01 GM115233/GM/NIGMS NIH HHS/United States
- DP5 OD012190/OD/NIH HHS/United States
- 1S10RR029591-01A1/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials