MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis - PubMed (original) (raw)
MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis
Alexander J Meeske et al. Proc Natl Acad Sci U S A. 2015.
Abstract
Bacterial surface polysaccharides are synthesized from lipid-linked precursors at the inner surface of the cytoplasmic membrane before being translocated across the bilayer for envelope assembly. Transport of the cell wall precursor lipid II in Escherichia coli requires the broadly conserved and essential multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily member MurJ. Here, we show that Bacillus subtilis cells lacking all 10 MOP superfamily members are viable with only minor morphological defects, arguing for the existence of an alternate lipid II flippase. To identify this factor, we screened for synthetic lethal partners of MOP family members using transposon sequencing. We discovered that an uncharacterized gene amj (alternate to MurJ; ydaH) and B. subtilis MurJ (murJBs; formerly ytgP) are a synthetic lethal pair. Cells defective for both Amj and MurJBs exhibit cell shape defects and lyse. Furthermore, expression of Amj or MurJBs in E. coli supports lipid II flipping and viability in the absence of E. coli MurJ. Amj is present in a subset of gram-negative and gram-positive bacteria and is the founding member of a novel family of flippases. Finally, we show that Amj is expressed under the control of the cell envelope stress-response transcription factor σ(M) and cells lacking MurJBs increase amj transcription. These findings raise the possibility that antagonists of the canonical MurJ flippase trigger expression of an alternate translocase that can resist inhibition.
Keywords: MurJ; flippase; lipid II; peptidoglycan; sigM.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
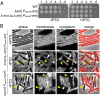
B. subtilis cells lacking all 10 MOP superfamily members are viable. (A) Growth curve of a strain lacking all 10 (∆10) MOP exporter superfamily members in B. subtilis. WT (BDR11) and the ∆10 mutant (BAM485) were grown to exponential phase in casein hydrolysate (CH) medium and back-diluted to an OD600 of 0.05, and growth was monitored over time. (B) Cytological analysis of the ∆10 mutant. Exponentially growing WT (BDR2648) and the ∆10 mutant (BAM486) were stained with the lipophilic dye 1-(4-trimethylammoniumphenyl)-6-phenyl-1,3,5-hexatriene _p_-toluenesulfonate and examined by fluorescence microscopy. Both strains express mCherry to label the cytoplasm. (Scale bar: 1 μm.)
Fig. 2.
Synthetic lethal screen using transposon sequencing identifies amj. Mariner transposon libraries were generated in WT (BDR2413) and the ∆4 mutant (BAM490) lacking ytgP, yabM, spoVB, and ykvU. The sites of transposon insertion were identified by deep sequencing and mapped onto the B. subtilis 168 reference genome (National Center for Biotechnology Information RefSeq NC_000964.3). Two regions of the genome are depicted. The height of each line reflects the number of sequencing reads at this position. Transposon insertions in sigM and amj (ydaH) were significantly reduced in the ∆4 mutant compared with WT. No insertions were mapped to the essential gene plsC in either strain. Despite the essentiality of yhdL, which encodes an anti-σM factor, some insertions were detected in the WT background.
Fig. 3.
MurJBs and Amj are synthetically lethal partners. (A) Spot dilutions of the indicated strains in the presence and absence of inducer. The three strains were grown in the presence of IPTG (500 μM) to an optical density of ∼1. The cultures were washed twice without inducer, resuspended at an OD600 of 1.25, and serially diluted. Five microliters of each dilution was spotted onto plates with and without inducer. (B) Cytological analysis of the terminal phenotype following Amj depletion in the absence of MurJBs. Depletion of the lipid II synthase MurG is shown for comparison. The indicated strains (BAM585 and BAM587) were grown to exponential phase in the presence of IPTG (500 μM), washed twice with media lacking inducer, back-diluted to an OD600 of 0.05 in CH medium, and grown to midexponential phase in the presence or absence of inducer. Cells were examined by fluorescence microscopy. Images of Amj depletion in the absence of MurJBs and MurG depletion are from 5 h and 4 h after removal of IPTG, respectively. Both strains contain mCherry, and an overlay of cytoplasmic fluorescence and phase contrast (merge) is shown. Lysis (yellow carets) and asymmetrical bulging (white carets) are highlighted. (Scale bar: 1 μm.)
Fig. 4.
Amj can substitute for MurJEc in E. coli. (A) Spot dilutions of E. coli strains in which expression of murJ_Ec was under the control of the arabinose-inducible (PBAD) promoter. The indicated genes were present on a medium copy plasmid and expressed from the P_lac promoter. Strains were grown to stationary phase in the presence of 0.2% arabinose, washed twice without inducer, resuspended at an OD600 of 1.25, and serially diluted. Five microliters of each dilution was spotted in the presence of 0.2% arabinose (+ARA) or 25 μM IPTG (−ARA). (B) Amj expression suppresses the loss of viability following depletion of MurJEc. Strains CAM252 (MurJEc depletion + pGFP) and CAM249 (MurJEc depletion + pAmj) were grown to stationary phase in the presence of arabinose, washed twice in LB lacking inducer, and back-diluted to an OD600 of 0.01 in LB containing arabinose (+ARA) or IPTG (−ARA). The cultures were grown for seven generations and back-diluted to an OD600 of 0.1, maintaining arabinose or IPTG. Cells were examined by differential interference contrast microscopy at hour 1.5 after the second back-dilution. Examples of bulging (white carets) and lysis (yellow carets) are highlighted. (Scale bar: 1 μm.) (C) Cytological analysis of E. coli strains lacking MurJEc complemented with P_lac_-amj or P_lac_-_murJ_Bs compared with WT. Strains were grown in LB containing 1 mM IPTG. (Scale bar: 1 μm.)
Fig. 5.
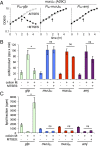
Amj can support lipid II flippase activity in E. coli. (A) Growth curves of E. coli murJ(A29C) ∆lysA strains producing GFP, MurJBs, or Amj treated with MTSES. Cultures were grown in minimal media with 1 mM IPTG until midexponential phase, followed by addition of MTSES as indicated (arrows). Growth was monitored over time. (B) In vivo flippase assay. Cells were labeled with [3H]-_m_-DAP. After 15 min, colicin M and/or MTSES was added as indicated, and growth was continued for 10 min. Samples were then withdrawn and extracted sequentially with hot water and then butanol. Hot-water extracts were subjected to HPLC and radiodetection to quantify the radiolabeled colicin M product. Experimental details and peak identification are provided in Fig. S7. (C) Scintillation counting was used to quantify label in the lipid (butanol) fraction. Error bars represent SEM from two experiments. The P value determined with Student’s t test. *P < 0.05. ns, not significant.
Fig. 6.
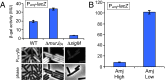
Amj expression increases in the absence of MurJBs. (A) Bar graph showing β-gal activity of indicated strains harboring the σM-responsive promoter (P_amj_) fused to lacZ. Activity was assayed in exponentially growing cultures in LB. Indicated strains harboring an amj promoter fusion to yfp were analyzed by fluorescence microscopy. (Scale bar: 1 μm.) (B) β-Gal activity of P_amj_-lacZ in a strain lacking _murJ_Bs with amj under an IPTG-inducible control. Activity was assayed in exponentially growing cultures in LB containing either 500 μM IPTG (Amj High) or 50 μM IPTG (Amj Low). Error bars represent SEM from two experiments.
Similar articles
- Bacillus subtilis MurJ and Amj Lipid II flippases are not essential for growth.
Englehart K, Dworkin J. Englehart K, et al. J Bacteriol. 2025 May 22;207(5):e0007825. doi: 10.1128/jb.00078-25. Epub 2025 Apr 4. J Bacteriol. 2025. PMID: 40183557 Free PMC article. - The O-Antigen Flippase Wzk Can Substitute for MurJ in Peptidoglycan Synthesis in Helicobacter pylori and Escherichia coli.
Elhenawy W, Davis RM, Fero J, Salama NR, Felman MF, Ruiz N. Elhenawy W, et al. PLoS One. 2016 Aug 18;11(8):e0161587. doi: 10.1371/journal.pone.0161587. eCollection 2016. PLoS One. 2016. PMID: 27537185 Free PMC article. - Loss of specificity variants of WzxC suggest that substrate recognition is coupled with transporter opening in MOP-family flippases.
Sham LT, Zheng S, Yakhnina AA, Kruse AC, Bernhardt TG. Sham LT, et al. Mol Microbiol. 2018 Sep;109(5):633-641. doi: 10.1111/mmi.14002. Epub 2018 Sep 15. Mol Microbiol. 2018. PMID: 29907971 Free PMC article. - Structure and Mechanism of the Lipid Flippase MurJ.
Kuk ACY, Hao A, Lee SY. Kuk ACY, et al. Annu Rev Biochem. 2022 Jun 21;91:705-729. doi: 10.1146/annurev-biochem-040320-105145. Epub 2022 Mar 23. Annu Rev Biochem. 2022. PMID: 35320686 Free PMC article. Review. - Lipid Flippases for Bacterial Peptidoglycan Biosynthesis.
Ruiz N. Ruiz N. Lipid Insights. 2016 Jan 13;8(Suppl 1):21-31. doi: 10.4137/LPI.S31783. eCollection 2015. Lipid Insights. 2016. PMID: 26792999 Free PMC article. Review.
Cited by
- Lipid somersaults: Uncovering the mechanisms of protein-mediated lipid flipping.
Pomorski TG, Menon AK. Pomorski TG, et al. Prog Lipid Res. 2016 Oct;64:69-84. doi: 10.1016/j.plipres.2016.08.003. Epub 2016 Aug 12. Prog Lipid Res. 2016. PMID: 27528189 Free PMC article. Review. - Bacterial developmental checkpoint that directly monitors cell surface morphogenesis.
Delerue T, Anantharaman V, Gilmore MC, Popham DL, Cava F, Aravind L, Ramamurthi KS. Delerue T, et al. Dev Cell. 2022 Feb 7;57(3):344-360.e6. doi: 10.1016/j.devcel.2021.12.021. Epub 2022 Jan 21. Dev Cell. 2022. PMID: 35065768 Free PMC article. - Genome-Wide Transposon Mutagenesis Screens Identify Group A Streptococcus Genes Affecting Susceptibility to β-Lactam Antibiotics.
Zhu L, Eraso JM, Mangham RE, Ojeda Saavedra M, Olsen RJ, Beres SB, Musser JM. Zhu L, et al. J Bacteriol. 2022 Dec 20;204(12):e0028722. doi: 10.1128/jb.00287-22. Epub 2022 Nov 14. J Bacteriol. 2022. PMID: 36374114 Free PMC article. - The Bacillus subtilis cell envelope stress-inducible ytpAB operon modulates membrane properties and contributes to bacitracin resistance.
Willdigg JR, Patel Y, Arquilevich BE, Subramanian C, Frank MW, Rock CO, Helmann JD. Willdigg JR, et al. J Bacteriol. 2024 Mar 21;206(3):e0001524. doi: 10.1128/jb.00015-24. Epub 2024 Feb 7. J Bacteriol. 2024. PMID: 38323910 Free PMC article. - Functional redundancy between penicillin-binding proteins during asymmetric cell division in Clostridioides difficile.
Shrestha S, Dressler JM, Harrison GA, McNellis ME, Shen A. Shrestha S, et al. bioRxiv [Preprint]. 2025 Mar 7:2024.09.26.615255. doi: 10.1101/2024.09.26.615255. bioRxiv. 2025. PMID: 39386573 Free PMC article. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
- GM086466/GM/NIGMS NIH HHS/United States
- RC2 GM092616/GM/NIGMS NIH HHS/United States
- R01 AI083365/AI/NIAID NIH HHS/United States
- GM102790/GM/NIGMS NIH HHS/United States
- AI083365-01/AI/NIAID NIH HHS/United States
- AI099144/AI/NIAID NIH HHS/United States
- R01 AI099144/AI/NIAID NIH HHS/United States
- R01 GM086466/GM/NIGMS NIH HHS/United States
- R01 GM102790/GM/NIGMS NIH HHS/United States
- U19 AI109764/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases