Integrating chemical mutagenesis and whole-genome sequencing as a platform for forward and reverse genetic analysis of Chlamydia - PubMed (original) (raw)
Integrating chemical mutagenesis and whole-genome sequencing as a platform for forward and reverse genetic analysis of Chlamydia
Marcela Kokes et al. Cell Host Microbe. 2015.
Abstract
Gene inactivation by transposon insertion or allelic exchange is a powerful approach to probe gene function. Unfortunately, many microbes, including Chlamydia, are not amenable to routine molecular genetic manipulations. Here we describe an arrayed library of chemically induced mutants of the genetically intransigent pathogen Chlamydia trachomatis, in which all mutations have been identified by whole-genome sequencing, providing a platform for reverse genetic applications. An analysis of possible loss-of-function mutations in the collection uncovered plasticity in the central metabolic properties of this obligate intracellular pathogen. We also describe the use of the library in a forward genetic screen that identified InaC as a bacterial factor that binds host ARF and 14-3-3 proteins and modulates F-actin assembly and Golgi redistribution around the pathogenic vacuole. This work provides a robust platform for reverse and forward genetic approaches in Chlamydia and should serve as a valuable resource to the community.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
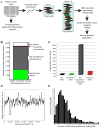
Figure 1. Generation of an ordered array of sequenced C. trachomatis mutants for use in genetic analyses
(A) Chemically mutagenized C. trachomatis (C. t) LGV-L2 434/Bu strains were clonally isolated by plaque purification and arrayed in 96 well plates. Genomic sequencing libraries from a pool of 20 mutant strains and 1 reference strain were tagged with a unique barcode, combined with four other pools, and sequenced on an Illumina NGS platform. (B) Distribution of synonymous (green), non synonymous (gray), nonsense (red), and non coding (white) single nucleotide variants (SNVs) identified among mutagenized C. t strains. (C) Distribution of ORFs without variant alleles (2%, black), with alleles harboring only synonymous SNVs (2%, green), containing both synonymous and non synonymous SNVs (96%, gray), and with nonsense SNVs (10%, red). (D) Density of SNVs (number of SNVs/10 k. b of DNA sequence) across the C. t LGV-L2 genome. * denotes rRNA paralogues (CTL_r01_2 CTL_r02_2). Underrepresentation of SNVs is due to an inability to unambiguously map sequencing reads to these loci. (E) Distribution of the number of variant alleles identified per ORF.
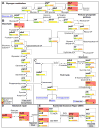
Figure 2. C. trachomatis tolerates perturbations in the tricarboxylic cycle and DNA repair networks
(A–C) The LGV C. t central carbon metabolic network. The distribution of all synonymous (green), neutral (yellow), non-neutral (orange), and nonsense (red) bearing alleles is depicted for each ORF in the network. Distributions are normalized to total number of SNV bearing alleles identified per ORF. All ORFs with observed nonsense alleles are shaded in light red. (A) Production and break down of branched glycogen is not necessary as LGV C. t tolerates nonsense mutations in glgB and glgX. (B) The presence of a nonsense mutation in pgi suggests that the pentose phosphate pathway is sufficient to circumvent loss of fructose-6-phosphate production when the EMP pathway (Glycolysis) is blocked. (C) Nonsense mutations in glt and sucA suggest that LGV biovars can sustain metabolic activity with a severely curtailed TCA cycle. The TCA cycle is further truncated due the presence of a fumC (dashed box) pseudogene. All compounds imported from the host are shown in blue. (D) LGV C. t biovars can tolerate defects in their DNA mismatch repair machinery as suggested by the presence of nonsense bearing alleles in mutS and mutL. (E) Defects in the nucleotide excision repair pathway are also tolerated as evidenced by the presence of nonsense bearing alleles in uvrA and uvrC.
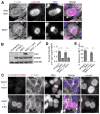
Figure 3. The inclusion membrane protein InaC is required for F-actin assembly at the inclusion
(A) F-actin assembly at inclusions (arrowhead) is absent in HeLa cells infected with M407. F-actin (grayscale) and C. t (red) were detected by indirect immunofluorescence at 30 hpi using rhodamine-phalloidin and anti-MOMP antibodies respectively. DNA (blue) was stained with Hoechst. (B–E) F-actin assembly in M407 inclusions is rescued upon expression of wild type InaC. InaC expression is restored in M407 mutants transformed with a plasmid expressing wild type inaC from its own promoter as assessed by immunoblot (C) and immunofluorescence (red, D) analysis and without affecting the distribution of another inclusion membrane protein, IncG. F-actin assembly at the inclusion (white arrowheads, (C)) is restored in M407 transformed with a plasmid encoding inaC but not an empty vector. Note that the average intensity (D) and frequency (E) of F-actin around complemented inclusions is comparable to wild type. Mean ± SEM for three independent experiments is shown. At least 50 (F) or 300 (E) inclusions were enumerated in each triplicate for each experiment. * indicates P < 0.05 by one-way ANOVA and Newman-Keuls post hoc. Scale bars represent 10μm.
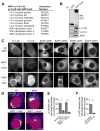
Figure 4. The recruitment of ARF and 14-3-3 proteins and Golgi redistribution to the inclusion requires InaC
(A and B) ARF and 14-3-3 proteins interact with InaC. Lysates from HEK 293T cells expressing either GFP or a GFP-InaC fusion protein were incubated with GFP-Trap resin. Bound proteins were identified by LC-MS/MS (Table S5). Proteins of ARF and 14-3-3 families are shown with corresponding accession numbers (A). The specificity of these interactions was confirmed by SDS-PAGE followed by western blot with specific antibodies (B). (C) InaC mediates the recruitment of 14-3-3β, -ε, and Golgi-specific ARF GTPases to the inclusion. HeLa cells were infected with the indicated C. t strains and either processed at 30 hpi for immunofluorescence using anti-14-3-3 antibodies (left) or transfected with EGFP-ARF isoforms for 24 hours before processing at 30 hpi for microscopy (right). Note that the intensity of 14-3-3 and ARF1, 4 and 5 recruitment to inclusions correlates with InaC expression. The divergent ARF6 was not recruited to inclusions or bound to GFP-InaC. (D and E) Golgi redistribution around the inclusion requires InaC. HeLa cells were infected as in (C) and processed at 30 hpi for immunofluorescence using anti-GM130 (grayscale) anti-IncG (red) antibodies and Hoechst (blue), and the frequency of Golgi distribution was quantified. Golgi distribution at least halfway around inclusions was scored as a positive (E), as detailed in Supplemental Experimental Procedures and Figure S3A. (F) F-actin assembly is required for Golgi redistribution at inclusions. HeLa cells were infected with a wild type LGV-L2 strain and treated with 500nM Latrunculin B for 30 minutes before processing as in (E) at 30 hpi. Mean ± SEM for three independent experiments is shown. At least 40 (E) or 90 (F) inclusions were enumerated in each triplicate for each experiment. * indicates P < 0.05 by one-way ANOVA and Newman-Keuls post hoc (E) or Student’s t test (F). Scale bars represent 10μm.
Similar articles
- Transposon Mutagenesis in Chlamydia trachomatis Identifies CT339 as a ComEC Homolog Important for DNA Uptake and Lateral Gene Transfer.
LaBrie SD, Dimond ZE, Harrison KS, Baid S, Wickstrum J, Suchland RJ, Hefty PS. LaBrie SD, et al. mBio. 2019 Aug 6;10(4):e01343-19. doi: 10.1128/mBio.01343-19. mBio. 2019. PMID: 31387908 Free PMC article. - Gene Deletion by Fluorescence-Reported Allelic Exchange Mutagenesis in Chlamydia trachomatis.
Mueller KE, Wolf K, Fields KA. Mueller KE, et al. mBio. 2016 Jan 19;7(1):e01817-15. doi: 10.1128/mBio.01817-15. mBio. 2016. PMID: 26787828 Free PMC article. - Forward and Reverse Genetic Analysis of Chlamydia.
Kędzior M, Bastidas RJ. Kędzior M, et al. Methods Mol Biol. 2019;2042:185-204. doi: 10.1007/978-1-4939-9694-0_13. Methods Mol Biol. 2019. PMID: 31385277 Free PMC article. - Advances in genetic manipulation of Chlamydia trachomatis.
Wan W, Li D, Li D, Jiao J. Wan W, et al. Front Immunol. 2023 Jun 28;14:1209879. doi: 10.3389/fimmu.2023.1209879. eCollection 2023. Front Immunol. 2023. PMID: 37449211 Free PMC article. Review. - [Information from the complete genome sequence of Chlamydia trachomatis: an obligate intracellular pathogen of humans].
Hagiwara T. Hagiwara T. Tanpakushitsu Kakusan Koso. 2000 Jun;45(8):1367-70. Tanpakushitsu Kakusan Koso. 2000. PMID: 10846474 Review. Japanese. No abstract available.
Cited by
- Transposon Mutagenesis in Chlamydia trachomatis Identifies CT339 as a ComEC Homolog Important for DNA Uptake and Lateral Gene Transfer.
LaBrie SD, Dimond ZE, Harrison KS, Baid S, Wickstrum J, Suchland RJ, Hefty PS. LaBrie SD, et al. mBio. 2019 Aug 6;10(4):e01343-19. doi: 10.1128/mBio.01343-19. mBio. 2019. PMID: 31387908 Free PMC article. - Bacterial genetics and molecular pathogenesis in the age of high throughput DNA sequencing.
Davey L, Valdivia RH. Davey L, et al. Curr Opin Microbiol. 2020 Apr;54:59-66. doi: 10.1016/j.mib.2020.01.007. Epub 2020 Feb 7. Curr Opin Microbiol. 2020. PMID: 32044689 Free PMC article. Review. - The cytoskeleton in cell-autonomous immunity: structural determinants of host defence.
Mostowy S, Shenoy AR. Mostowy S, et al. Nat Rev Immunol. 2015 Sep 15;15(9):559-73. doi: 10.1038/nri3877. Epub 2015 Aug 21. Nat Rev Immunol. 2015. PMID: 26292640 Free PMC article. Review. - Chlamydia Hijacks ARF GTPases To Coordinate Microtubule Posttranslational Modifications and Golgi Complex Positioning.
Wesolowski J, Weber MM, Nawrotek A, Dooley CA, Calderon M, St Croix CM, Hackstadt T, Cherfils J, Paumet F. Wesolowski J, et al. mBio. 2017 May 2;8(3):e02280-16. doi: 10.1128/mBio.02280-16. mBio. 2017. PMID: 28465429 Free PMC article. - IncV, a FFAT motif-containing Chlamydia protein, tethers the endoplasmic reticulum to the pathogen-containing vacuole.
Murray R, Flora E, Bayne C, Derré I. Murray R, et al. Proc Natl Acad Sci U S A. 2017 Nov 7;114(45):12039-12044. doi: 10.1073/pnas.1709060114. Epub 2017 Oct 23. Proc Natl Acad Sci U S A. 2017. PMID: 29078338 Free PMC article.
References
- Beagley KW, Huston WM, Hansbro PM, Timms P. Chlamydial infection of immune cells: altered function and implications for disease. Crit Rev Immunol. 2009;29:275–305. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R56 AI107951/AI/NIAID NIH HHS/United States
- AI102500-01/AI/NIAID NIH HHS/United States
- AI081694/AI/NIAID NIH HHS/United States
- F32 AI102500/AI/NIAID NIH HHS/United States
- AI100759/AI/NIAID NIH HHS/United States
- AI085238/AI/NIAID NIH HHS/United States
- R01 AI100759/AI/NIAID NIH HHS/United States
- R01 AI107951/AI/NIAID NIH HHS/United States
- R21 AI085238/AI/NIAID NIH HHS/United States
- R01 AI081694/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources