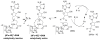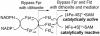Chemical and Biological Reduction of the Radical SAM Enzyme 7-Carboxy-7-deazaguanine [corrected] Synthase - PubMed (original) (raw)
Chemical and Biological Reduction of the Radical SAM Enzyme 7-Carboxy-7-deazaguanine [corrected] Synthase
Nathan A Bruender et al. Biochemistry. 2015.
Erratum in
- Correction to Chemical and Biological Reduction of the Radical SAM Enzyme 7-Carboxy-7-deazaguanine Synthase.
Bruender NA, Young AP, Bandarian V. Bruender NA, et al. Biochemistry. 2016 Mar 29;55(12):1939. doi: 10.1021/acs.biochem.6b00147. Epub 2016 Mar 7. Biochemistry. 2016. PMID: 26950390 No abstract available.
Abstract
The radical S-adenosyl-L-methionine (SAM) superfamily is a large and growing group of enzymes that conduct complex radical-mediated transformations. A one-electron reduction of SAM via the +1 state of the cubane [4Fe-4S] cluster generates a 5'-deoxyadenosyl radical, which initiates turnover. The [4Fe-4S] cluster must be reduced from its resting +2 state to the catalytically active +1 oxidation state by an electron. In practice, dithionite or the Escherichia coli flavodoxin (EcFldA)/ferredoxin (flavodoxin):NADP(+) oxidoreductase (Fpr)/NADPH system is used. Herein, we present a systematic investigation of the reductive activation of the radical SAM enzyme CDG synthase (BsQueE) from Bacillus subtilis comparing biological and chemical reductants. These data show that either of the flavodoxin homologues encoded by the B. subtilis genome, BsYkuN or BsYkuP, as well as a series of small molecule redox mediators, supports BsQueE activity. With dithionite as a reductant, the activity of BsQueE is ~75-fold greater in the presence of BsYkuN and BsYkuP compared to that in the presence of dithionite alone. By contrast, EcFldA supports turnover to ~10-fold greater levels than dithionite alone under the same conditions. Comparing the ratio of the rate of turnover to the apparent binding constant for the flavodoxin homologues reveals 10- and 240-fold preferences for BsYkuN over BsYkuP and EcFldA, respectively. The differential activation of the enzyme cannot be explained by the abortive cleavage of SAM. We conclude from these observations that the differential activation of BsQueE by Fld homologues may reside in the details of the interaction between the flavodoxin and the radical SAM enzyme.
Figures
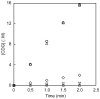
Figure 1
Time course of _Bs_QueE reaction under various reducing conditions. Activity was monitored by measuring CDG formation._Bs_QueE activity was measured in the presence 10 mM dithionite (upside-down triangle) or 2 μM Fpr/2 mM NADPH with 5 μM_Bs_YkuN (circle), 5 μM Fpr/2 mM NADPH with 20 μM _Bs_YkuP (triangle), or 5 μM Fpr/2 mM NADPH with 20 μM _Ec_FldA (diamond). Assays where Fpr or NADPH (square) were omitted served as negative controls.
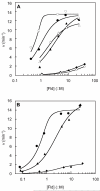
Figure 2
_Bs_QueE activity is enhanced by increasing the flavodoxin homologs [_Ec_FldA(diamonds)], [_Bs_YkuN (circles)], or [_Bs_YkuP (triangles)] concentration when the electrons are supplied by either NADPH via Fpr (A) or dithionite (B). To confirm Fld reduction by NADPH/Fpr was saturating, the rate of CDG formation dependent on [Fld] was measured at 1 μM (closed) and 5 μM (open) Fpr for _Bs_YkuN and 5 μM (closed) and 20 μM (open) for _Bs_YkuP and _Ec_FldA (A). Depicted are representative data sets for each experiment. The data were fit using Eqn 1 and the _V_max (_k_CDG) ant_K_Fld are reported in Table 1.
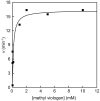
Figure 3
_Bs_QueE activity is enhanced by increasing methyl viologen concentration when electrons are supplied by dithionite. The data were fit using Eqn. 2 to determine_k_CDG. The activity of _Bs_QueE was 0.2 min−1 in the absence of methyl viologen.
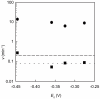
Figure 4
Velocity of CDG formation (circles) or dAdo formation (squares) as a function of reduction potential of redox mediator used in the activity assay. Dithionite was the reductant used in all of the reactions. The redox mediators used were methyl viologen (Em= −0.446 V), benzyl viologen (Em= −0.359 V), neutral red (Em= −0.325 V), and lissamine blue BF (Em= −0.276 V, pH 7.5). The dashed lines represent the rates of formation for CDG (–––) and dAdo (---), 0.2 min−1 and 0.08 min−1respectively, when _Bs_QueE was activated by dithionite alone.
Scheme 1
Reductive cleavage of SAM.
Scheme 2
Electron flow for [4Fe-4S] cluster reduction.a a The scheme shows the flow of electrons and does not imply absolute stoichiometry.
Similar articles
- 7-Carboxy-7-deazaguanine Synthase: A Radical S-Adenosyl-l-methionine Enzyme with Polar Tendencies.
Bruender NA, Grell TA, Dowling DP, McCarty RM, Drennan CL, Bandarian V. Bruender NA, et al. J Am Chem Soc. 2017 Feb 8;139(5):1912-1920. doi: 10.1021/jacs.6b11381. Epub 2017 Jan 25. J Am Chem Soc. 2017. PMID: 28045519 Free PMC article. - Activation of class III ribonucleotide reductase by flavodoxin: a protein radical-driven electron transfer to the iron-sulfur center.
Mulliez E, Padovani D, Atta M, Alcouffe C, Fontecave M. Mulliez E, et al. Biochemistry. 2001 Mar 27;40(12):3730-6. doi: 10.1021/bi001746c. Biochemistry. 2001. PMID: 11297442 - A Radical Intermediate in Bacillus subtilis QueE during Turnover with the Substrate Analogue 6-Carboxypterin.
Wilcoxen J, Bruender NA, Bandarian V, Britt RD. Wilcoxen J, et al. J Am Chem Soc. 2018 Feb 7;140(5):1753-1759. doi: 10.1021/jacs.7b10860. Epub 2018 Jan 24. J Am Chem Soc. 2018. PMID: 29303575 Free PMC article. - Emerging themes in radical SAM chemistry.
Shisler KA, Broderick JB. Shisler KA, et al. Curr Opin Struct Biol. 2012 Dec;22(6):701-10. doi: 10.1016/j.sbi.2012.10.005. Epub 2012 Nov 8. Curr Opin Struct Biol. 2012. PMID: 23141873 Free PMC article. Review. - Auxiliary iron-sulfur cofactors in radical SAM enzymes.
Lanz ND, Booker SJ. Lanz ND, et al. Biochim Biophys Acta. 2015 Jun;1853(6):1316-34. doi: 10.1016/j.bbamcr.2015.01.002. Epub 2015 Jan 15. Biochim Biophys Acta. 2015. PMID: 25597998 Review.
Cited by
- 1,2-Propanediol Dehydration in Roseburia inulinivorans: STRUCTURAL BASIS FOR SUBSTRATE AND ENANTIOMER SELECTIVITY.
LaMattina JW, Keul ND, Reitzer P, Kapoor S, Galzerani F, Koch DJ, Gouvea IE, Lanzilotta WN. LaMattina JW, et al. J Biol Chem. 2016 Jul 22;291(30):15515-26. doi: 10.1074/jbc.M116.721142. Epub 2016 Jun 1. J Biol Chem. 2016. PMID: 27252380 Free PMC article. - Biosynthesis and function of 7-deazaguanine derivatives in bacteria and phages.
de Crécy-Lagard V, Hutinet G, Cediel-Becerra JDD, Yuan Y, Zallot R, Chevrette MG, Ratnayake RMMN, Jaroch M, Quaiyum S, Bruner S. de Crécy-Lagard V, et al. Microbiol Mol Biol Rev. 2024 Mar 27;88(1):e0019923. doi: 10.1128/mmbr.00199-23. Epub 2024 Feb 29. Microbiol Mol Biol Rev. 2024. PMID: 38421302 Review. - QueE: A Radical SAM Enzyme Involved in the Biosynthesis of 7-Deazapurine Containing Natural Products.
Lewis JK, Bruender NA, Bandarian V. Lewis JK, et al. Methods Enzymol. 2018;606:95-118. doi: 10.1016/bs.mie.2018.05.001. Epub 2018 Jul 13. Methods Enzymol. 2018. PMID: 30097106 Free PMC article. - Journey on the Radical SAM Road as an Accidental Pilgrim.
Bandarian V. Bandarian V. ACS Bio Med Chem Au. 2022 Jun 15;2(3):187-195. doi: 10.1021/acsbiomedchemau.1c00059. Epub 2022 Feb 28. ACS Bio Med Chem Au. 2022. PMID: 35726327 Free PMC article. Review. - Eukaryotic TYW1 Is a Radical SAM Flavoenzyme.
Young AP, Bandarian V. Young AP, et al. Biochemistry. 2021 Jul 13;60(27):2179-2185. doi: 10.1021/acs.biochem.1c00349. Epub 2021 Jun 29. Biochemistry. 2021. PMID: 34184886 Free PMC article.
References
- Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a Novel Protein Superfamily Linking Unresolved Steps In Familiar Biosynthetic Pathways With Radical Mechanisms: Functional Characterization Using New Analysis And Information Visualization Methods. Nucleic Acids Res. 2001;29:1097–1106. - PMC - PubMed
- Walsby CJ, Ortillo D, Broderick WE, Broderick JB, Hoffman BM. An Anchoring Role for FeS Clusters: Chelation of the Amino Acid Moiety of S-Adenosylmethionine to the Unique Iron Site of the [4Fe–4S] Cluster of Pyruvate Formate-Lyase Activating Enzyme. J. Am. Chem. Soc. 2002;124:11270–11271. - PubMed
- Chen D, Walsby C, Hoffman BM, Frey PA. Coordination and Mechanism of Reversible Cleavage of S-Adenosylmethionine by the [4Fe-4S] Center in Lysine 2,3-Aminomutase. J. Am. Chem. Soc. 2003;125:11788–11789. - PubMed
- Knappe J, Schacht J, Mockel W, Hopner T, Vetter H, Edenharder R. Pyruvate Formate-Lyase Reaction in Escherichia coli. The Enzymatic System Converting an Inactive Form of the Lyase into the Catalytically Active Enzyme. Eur J Biochem. 1969;11:316–327. - PubMed
- Chirpich TP, Zappia V, Costilow RN, Barker HA. Lysine 2, 3-Aminomutase. Purification And Properties Of A Pyridoxal Phosphate And S-Adenosylmethionine-Activated Enzyme. J. Biol. Chem. 1970;245:1778–1789. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases