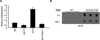DNA Methylation on N6-Adenine in C. elegans - PubMed (original) (raw)
DNA Methylation on N6-Adenine in C. elegans
Eric Lieberman Greer et al. Cell. 2015.
Abstract
In mammalian cells, DNA methylation on the fifth position of cytosine (5mC) plays an important role as an epigenetic mark. However, DNA methylation was considered to be absent in C. elegans because of the lack of detectable 5mC, as well as homologs of the cytosine DNA methyltransferases. Here, using multiple approaches, we demonstrate the presence of adenine N(6)-methylation (6mA) in C. elegans DNA. We further demonstrate that this modification increases trans-generationally in a paradigm of epigenetic inheritance. Importantly, we identify a DNA demethylase, NMAD-1, and a potential DNA methyltransferase, DAMT-1, which regulate 6mA levels and crosstalk between methylations of histone H3K4 and adenines and control the epigenetic inheritance of phenotypes associated with the loss of the H3K4me2 demethylase spr-5. Together, these data identify a DNA modification in C. elegans and raise the exciting possibility that 6mA may be a carrier of heritable epigenetic information in eukaryotes.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
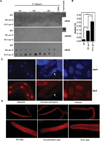
Figure 1. 6mA occurs in C. elegans DNA and increases across spr-5 generations
A) Dot blots of 3 biological replicates of WT, generation 5, and generation 15 spr-5(by101) mutant worms grown at 16°, 20°, or 25° all show progressively elevated 6mA and lack detectable 5mC and 5hmC. 250 ng of gDNA are loaded per dot. Mammalian gDNA is used as a control for 5mC and 5hmC antibody strength. B) 6mA levels increase across generations of spr-5(by101) mutant worms as assessed by UHPLC-MS/MS. Each column represents the mean and standard deviation of 3–5 biological replicates per group. * p < 0.05, **** p <0.0001. C) Immunofluorescence displays 6mA staining in the intestine, oocytes, and every cell of the embryo. Arrow indicates sperm. D) Immunofluorescence of wildtype extracted germlines show 6mA in every nuclei. This staining was competed by a 6mA premethylated oligo but not by unmethylated oligos. See also Figures S1, S2, and S3.
Figure 2. 6mA genomic location
A) Representative interpulse duration (IPD) ratios, of SMRT sequencing data of mixed stage wildtype worms. IPD ratio is defined as the change in IPD distribution in the sample compared to unmodified bases. Red: positive strand; Blue: negative strand. B) Comparison of observed vs. simulated distributions of 6mA across the C. elegans genome indicates that 6mA is not enriched or depleted in any major genomic feature. A permutation was used to calculate the average of 10,000 simulations for comparison to the observed data. C) Circos plots of 6mA and motif distributions; three inner rings: 6mA density normalized to adenines in each bin of 6mAs within different methylation fractions. Red, yellow, and blue represent highly methylated (80–100%), intermediate (20–80%), and lowly methylated (10–20%) 6mA, respectively. The middle ring shows AGAA (red) and GAGG (blue) motif densities, with purple indicating the overlap. The outer ring (rainfall plot) shows the distribution of inter-distance between each two adjacent 6mAs in the same motif. Red dots represent 6mAs in AGAA motif and blue dots represent 6mAs in GAGG motif; increasing vertical distance towards the center of the circle indicates increasing local density of 6mA occurrences. D) SMRT sequencing identified two motifs associated with 6mA. AGAA and GAGG are associated with low and high percentage 6mA, respectively. Methylation level refers to the percentage of times (1.0 = 100%) a given A in the sample population was read as methylated by SMRT sequencing. See also Figure S5 for 6mA MeDIPseq.
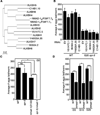
Figure 3. Deletion of nmad-1 accelerates the progressive fertility defect of spr-5 mutant worms
A) Phylogeny tree of human and C. elegans ALKBH family members. B) Knockdown of 4 of the ALKBH family members has no effect on egg laying of WT and spr-5 mutant worms treated for 20 generations with bacteria expressing the specific dsRNAs. Knockdown efficiency was tested by real-time RT PCR (Figure S6A). C) Early generation (G5) spr-5 mutant worms do not display significant fertility defects, but when combined with nmad-1 deletion these worms become sterile by generation 4. Each bar represents the mean ± SEM of 3 independent experiments. D) nmad-1 mutants lay fewer eggs than WT worms but do not display a progressive fertility decline. Each bar represents the mean ± SEM of 2–6 independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, ns not significant.
Figure 4. NMAD-1 demethylates 6mA in vitro and in vivo
A) Two different isoforms of NMAD-1 demethylate single stranded denatured (ss) and double stranded non-denatured (ds, hemi or dual methylated) oligos premethylated at 6mA and 3mC but not 1mA. B) Mutation of the catalytic domain of NMAD-1 abrogates the ability of NMAD-1 to demethylate 6mA premethylated oligos. C) nmad-1 mutants have elevated levels of 6mA without detectable changes in 3mC levels. Each dot represents 250 ng of DNA of independent biological replicates. D) nmad-1 mutants have elevated levels of 6mA as assessed by UHPLC-MS/MS. Each bar represents the mean and standard error of the mean of 2 independent biological replicates measured in duplicate. * p < 0.05.
Figure 5. DAMT-1 regulates 6mA levels
A) Phylogeny tree shows conservation of DAMT-1 in other eukaryotic species. Full tree and details of related clades are presented in Figure S6B. B) gDNA extracted from SF9 cells infected with full length or the catalytic domain of damt-1 show elevated levels of 6mA by dot blot. C) Mutation of the catalytic domain of DAMT-1 (DPPW to APPA) limits the increase in 6mA levels of infected SF9 cells. DAMT-1 expression is presented in Figure S6C. D) damt-1 knockdown decreases 6mA without affecting detectable 3mC levels. E) damt-1 mutants have decreased levels of 6mA as assessed by LC-MS/MS. Each bar represents the mean and standard error of the mean of 3 independent experiments of 3 biological replicates each measured in duplicate. ** p < 0.01. F) Generation 20 (G20) spr-5 mutant worms show elevated 6mA levels compared with WT worms and damt-1 knockdown suppresses the elevated 6mA in spr-5 mutant worms. Each dot represents 250 ng of gDNA of independent biological replicates.
Figure 6. Deletion of damt-1 suppresses the trans-generational phenotypes of spr-5 mutant worms
A) damt-1 knockdown has no effect on WT egg laying but partially suppresses the progressive fertility defect of spr-5(by101) mutant worms. B) damt-1 deletion has no effect on WT egg laying but partially suppresses the progressive fertility defect of spr-5(by134) mutant worms. C) damt-1 knockdown reverts the egg laying defect of nmad-1 mutant worms. All assays were performed at generation 20. Each bar represents the mean ± SEM of 3 independent experiments. * p < 0.05, ** p < 0.01, ns not significant.
Figure 7. DNA methylation and histone methylation crosstalk
A) Deletion of damt-1 suppresses the elevated H3K4me2 levels of late generation spr-5(by134) mutant worms. Each bar represents the mean ± SEM of 3 independent experiments performed in biological duplicate. Image J was used to analyze the relative intensity of H3K4me2 compared to Histone H3. Western blots corresponding to two of these experiments are shown in Figures S7A and S7B. B) Knockdown of H3K9me binding protein, eap-1, suppresses the elevated 6mA level detected in spr-5 mutant worms as assessed by dot blots. A longer exposure showing 6mA levels in WT worms is shown in Figure S7C.
Similar articles
- A histone methylation network regulates transgenerational epigenetic memory in C. elegans.
Greer EL, Beese-Sims SE, Brookes E, Spadafora R, Zhu Y, Rothbart SB, Aristizábal-Corrales D, Chen S, Badeaux AI, Jin Q, Wang W, Strahl BD, Colaiácovo MP, Shi Y. Greer EL, et al. Cell Rep. 2014 Apr 10;7(1):113-26. doi: 10.1016/j.celrep.2014.02.044. Epub 2014 Mar 27. Cell Rep. 2014. PMID: 24685137 Free PMC article. - DNA N(6)-methyladenine: a new epigenetic mark in eukaryotes?
Luo GZ, Blanco MA, Greer EL, He C, Shi Y. Luo GZ, et al. Nat Rev Mol Cell Biol. 2015 Dec;16(12):705-10. doi: 10.1038/nrm4076. Epub 2015 Oct 28. Nat Rev Mol Cell Biol. 2015. PMID: 26507168 Free PMC article. Review. - Fanconi Anemia FANCM/FNCM-1 and FANCD2/FCD-2 Are Required for Maintaining Histone Methylation Levels and Interact with the Histone Demethylase LSD1/SPR-5 in Caenorhabditis elegans.
Kim HM, Beese-Sims SE, Colaiácovo MP. Kim HM, et al. Genetics. 2018 Jun;209(2):409-423. doi: 10.1534/genetics.118.300823. Epub 2018 Mar 27. Genetics. 2018. PMID: 29588287 Free PMC article. - N6-Methyladenine DNA Modification in the Human Genome.
Xiao CL, Zhu S, He M, Chen D, Zhang Q, Chen Y, Yu G, Liu J, Xie SQ, Luo F, Liang Z, Wang DP, Bo XC, Gu XF, Wang K, Yan GR. Xiao CL, et al. Mol Cell. 2018 Jul 19;71(2):306-318.e7. doi: 10.1016/j.molcel.2018.06.015. Epub 2018 Jul 12. Mol Cell. 2018. PMID: 30017583 - Epigenetics in C. elegans: facts and challenges.
Wenzel D, Palladino F, Jedrusik-Bode M. Wenzel D, et al. Genesis. 2011 Aug;49(8):647-61. doi: 10.1002/dvg.20762. Epub 2011 Jul 22. Genesis. 2011. PMID: 21538806 Review.
Cited by
- Selective Effect of DNA N6-Methyladenosine Modification on Transcriptional Genetic Variations in East Asian Samples.
Luan M, Chen K, Zhao W, Tang M, Wang L, Liu S, Zhu L, Xie S. Luan M, et al. Int J Mol Sci. 2024 Sep 27;25(19):10400. doi: 10.3390/ijms251910400. Int J Mol Sci. 2024. PMID: 39408729 Free PMC article. - Dealkylation of Macromolecules by Eukaryotic α-Ketoglutarate-Dependent Dioxygenases from the AlkB-like Family.
Davletgildeeva AT, Kuznetsov NA. Davletgildeeva AT, et al. Curr Issues Mol Biol. 2024 Sep 20;46(9):10462-10491. doi: 10.3390/cimb46090622. Curr Issues Mol Biol. 2024. PMID: 39329974 Free PMC article. Review. - The biological function of demethylase ALKBH1 and its role in human diseases.
Zhong J, Xu Z, Ding N, Wang Y, Chen W. Zhong J, et al. Heliyon. 2024 Jun 24;10(13):e33489. doi: 10.1016/j.heliyon.2024.e33489. eCollection 2024 Jul 15. Heliyon. 2024. PMID: 39040364 Free PMC article. Review. - Symmetric and asymmetric DNA N6-adenine methylation regulates different biological responses in Mucorales.
Lax C, Mondo SJ, Osorio-Concepción M, Muszewska A, Corrochano-Luque M, Gutiérrez G, Riley R, Lipzen A, Guo J, Hundley H, Amirebrahimi M, Ng V, Lorenzo-Gutiérrez D, Binder U, Yang J, Song Y, Cánovas D, Navarro E, Freitag M, Gabaldón T, Grigoriev IV, Corrochano LM, Nicolás FE, Garre V. Lax C, et al. Nat Commun. 2024 Jul 18;15(1):6066. doi: 10.1038/s41467-024-50365-2. Nat Commun. 2024. PMID: 39025853 Free PMC article. - Adenosine Deaminase-Like Gene-Carried Lentivirus Toolkit for Identification of DNA N6-Methyladenine Origins.
Liang Z, Chen S, Li Y, Lai W, Wang H. Liang Z, et al. Adv Sci (Weinh). 2024 Sep;11(35):e2403376. doi: 10.1002/advs.202403376. Epub 2024 Jul 18. Adv Sci (Weinh). 2024. PMID: 39023073 Free PMC article.
References
- Arber W, Dussoix D. Host specificity of DNA produced by Escherichia coli. I. Host controlled modification of bacteriophage lambda. J Mol Biol. 1962;5:18–36. - PubMed
- Benyshek DC, Johnston CS, Martin JF. Glucose metabolism is altered in the adequately-nourished grand-offspring (F3 generation) of rats malnourished during gestation and perinatal life. Diabetologia. 2006;49:1117–1119. - PubMed
- Braun RE, Wright A. DNA methylation differentially enhances the expression of one of the two E. coli dnaA promoters in vivo and in vitro. Mol Gen Genet. 1986;202:246–250. - PubMed
- Cavalli G, Paro R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell. 1998;93:505–518. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 1F32CA180450-01/CA/NCI NIH HHS/United States
- P40 OD010440/OD/NIH HHS/United States
- R00 AG043550/AG/NIA NIH HHS/United States
- R01 CA118487/CA/NCI NIH HHS/United States
- K99 AG043550/AG/NIA NIH HHS/United States
- Intramural NIH HHS/United States
- MH096066/MH/NIMH NIH HHS/United States
- R01 MH096066/MH/NIMH NIH HHS/United States
- CA118487/CA/NCI NIH HHS/United States
- K99AG043550/AG/NIA NIH HHS/United States
- R01 GM058012/GM/NIGMS NIH HHS/United States
- GM058012/GM/NIGMS NIH HHS/United States
- Howard Hughes Medical Institute/United States
- F32 CA180450/CA/NCI NIH HHS/United States
- P40OD010440/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials