Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data - PubMed (original) (raw)
Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data
Kathrin P Aßhauer et al. Bioinformatics. 2015.
Abstract
Motivation: The characterization of phylogenetic and functional diversity is a key element in the analysis of microbial communities. Amplicon-based sequencing of marker genes, such as 16S rRNA, is a powerful tool for assessing and comparing the structure of microbial communities at a high phylogenetic resolution. Because 16S rRNA sequencing is more cost-effective than whole metagenome shotgun sequencing, marker gene analysis is frequently used for broad studies that involve a large number of different samples. However, in comparison to shotgun sequencing approaches, insights into the functional capabilities of the community get lost when restricting the analysis to taxonomic assignment of 16S rRNA data.
Results: Tax4Fun is a software package that predicts the functional capabilities of microbial communities based on 16S rRNA datasets. We evaluated Tax4Fun on a range of paired metagenome/16S rRNA datasets to assess its performance. Our results indicate that Tax4Fun provides a good approximation to functional profiles obtained from metagenomic shotgun sequencing approaches.
Availability and implementation: Tax4Fun is an open-source R package and applicable to output as obtained from the SILVAngs web server or the application of QIIME with a SILVA database extension. Tax4Fun is freely available for download at http://tax4fun.gobics.de/.
Contact: kasshau@gwdg.de
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2015. Published by Oxford University Press.
Figures
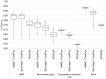
Fig. 1.
Spearman correlations between metagenomic and 16S-predicted functional profiles for comparison of Tax4Fun and PICRUSt on paired datasets from the human microbiome (HMP), mammalian guts, Guerrero Negro hypersaline microbial mat and soils
Similar articles
- Comparing bacterial communities inferred from 16S rRNA gene sequencing and shotgun metagenomics.
Shah N, Tang H, Doak TG, Ye Y. Shah N, et al. Pac Symp Biocomput. 2011:165-76. doi: 10.1142/9789814335058_0018. Pac Symp Biocomput. 2011. PMID: 21121044 - Inference-based accuracy of metagenome prediction tools varies across sample types and functional categories.
Sun S, Jones RB, Fodor AA. Sun S, et al. Microbiome. 2020 Apr 2;8(1):46. doi: 10.1186/s40168-020-00815-y. Microbiome. 2020. PMID: 32241293 Free PMC article. - VITCOMIC2: visualization tool for the phylogenetic composition of microbial communities based on 16S rRNA gene amplicons and metagenomic shotgun sequencing.
Mori H, Maruyama T, Yano M, Yamada T, Kurokawa K. Mori H, et al. BMC Syst Biol. 2018 Mar 19;12(Suppl 2):30. doi: 10.1186/s12918-018-0545-2. BMC Syst Biol. 2018. PMID: 29560821 Free PMC article. - Bioinformatics for NGS-based metagenomics and the application to biogas research.
Jünemann S, Kleinbölting N, Jaenicke S, Henke C, Hassa J, Nelkner J, Stolze Y, Albaum SP, Schlüter A, Goesmann A, Sczyrba A, Stoye J. Jünemann S, et al. J Biotechnol. 2017 Nov 10;261:10-23. doi: 10.1016/j.jbiotec.2017.08.012. Epub 2017 Aug 18. J Biotechnol. 2017. PMID: 28823476 Review. - [A review on the bioinformatics pipelines for metagenomic research].
Ye DD, Fan MM, Guan Q, Chen HJ, Ma ZS. Ye DD, et al. Dongwuxue Yanjiu. 2012 Dec;33(6):574-85. doi: 10.3724/SP.J.1141.2012.06574. Dongwuxue Yanjiu. 2012. PMID: 23266976 Review. Chinese.
Cited by
- Gut dysbiosis and clinical phases of pancolitis in patients with ulcerative colitis.
Maldonado-Arriaga B, Sandoval-Jiménez S, Rodríguez-Silverio J, Lizeth Alcaráz-Estrada S, Cortés-Espinosa T, Pérez-Cabeza de Vaca R, Licona-Cassani C, Gámez-Valdez JS, Shaw J, Mondragón-Terán P, Hernández-Cortez C, Suárez-Cuenca JA, Castro-Escarpulli G. Maldonado-Arriaga B, et al. Microbiologyopen. 2021 Mar;10(2):e1181. doi: 10.1002/mbo3.1181. Microbiologyopen. 2021. PMID: 33970546 Free PMC article. - Driving forces of soil bacterial community structure, diversity, and function in temperate grasslands and forests.
Kaiser K, Wemheuer B, Korolkow V, Wemheuer F, Nacke H, Schöning I, Schrumpf M, Daniel R. Kaiser K, et al. Sci Rep. 2016 Sep 21;6:33696. doi: 10.1038/srep33696. Sci Rep. 2016. PMID: 27650273 Free PMC article. - Comparison of the composition and function of the gut microbiome in herdsmen from two pasture regions, Hongyuan and Xilingol.
Yang C, Peng C, Jin H, You L, Wang J, Xu H, Sun Z. Yang C, et al. Food Sci Nutr. 2021 May 4;9(6):3258-3268. doi: 10.1002/fsn3.2290. eCollection 2021 Jun. Food Sci Nutr. 2021. PMID: 34136190 Free PMC article. - Gut dysbacteriosis induces expression differences in the adult head transcriptome of Spodoptera frugiperda in a sex-specific manner.
Junrui-Fu, Rong Z, Huang X, Wang J, Long X, Feng Q, Deng H. Junrui-Fu, et al. BMC Microbiol. 2023 Dec 7;23(1):388. doi: 10.1186/s12866-023-03089-0. BMC Microbiol. 2023. PMID: 38057708 Free PMC article. - Bacterial diversity of an acid mine drainage beside the Xichú River (Mexico) accessed by culture-dependent and culture-independent approaches.
Brito EMS, Guyoneaud R, Caretta CA, Joseph M, Goñi-Urriza M, Ollivier B, Hirschler-Réa A. Brito EMS, et al. Extremophiles. 2023 Feb 17;27(1):5. doi: 10.1007/s00792-023-01291-6. Extremophiles. 2023. PMID: 36800123
References
- Aßhauer K.P., Meinicke P. (2013) On the estimation of metabolic profiles in metagenomics. In: Beißbarth T., Kollmar M., Leha A., Morgenstern B., Schultz A.-K., Waack S., Wingender E. (eds.) German Conference on Bioinformatics 2013, volume 34 of OpenAccess Series in Informatics (OASIcs). Schloss Dagstuhl–Leibniz-Zentrum fuer Informatik: Dagstuhl, Germany, pp. 1–13.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources