Accessing DNA damage in chromatin: Preparing the chromatin landscape for base excision repair - PubMed (original) (raw)
Review
Accessing DNA damage in chromatin: Preparing the chromatin landscape for base excision repair
Yesenia Rodriguez et al. DNA Repair (Amst). 2015 Aug.
Abstract
DNA damage in chromatin comes in many forms, including single base lesions that induce base excision repair (BER). We and others have shown that the structural location of DNA lesions within nucleosomes greatly influences their accessibility to repair enzymes. Indeed, a difference in the location of uracil as small as one-half turn of the DNA backbone on the histone surface can result in a 10-fold difference in the time course of its removal in vitro. In addition, the cell has evolved several interdependent processes capable of enhancing the accessibility of excision repair enzymes to DNA lesions in nucleosomes, including post-translational modification of histones, ATP-dependent chromatin remodeling and interchange of histone variants in nucleosomes. In this review, we focus on different factors that affect accessibility of BER enzymes to nucleosomal DNA.
Keywords: AP endonuclease; Chromatin; Chromatin remodeling; Glycosylase; Histone acetylation; Histone variants; Ligase; Nucleosome dynamics; Polymerase β; Rotational setting.
Copyright © 2015 Elsevier B.V. All rights reserved.
Figures
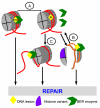
Figure 1
Schematic illustration of the various modalities that influence DNA accessibility to BER enzymes in NCPs: (A) Rotational orientation, (B) Intrinsic nucleosome dynamics and stability, and (C) ATP-dependent chromatin remodeling.
Similar articles
- Site-specific Acetylation of Histone H3 Decreases Polymerase β Activity on Nucleosome Core Particles in Vitro.
Rodriguez Y, Hinz JM, Laughery MF, Wyrick JJ, Smerdon MJ. Rodriguez Y, et al. J Biol Chem. 2016 May 20;291(21):11434-45. doi: 10.1074/jbc.M116.725788. Epub 2016 Mar 31. J Biol Chem. 2016. PMID: 27033702 Free PMC article. - A cassette of basic amino acids in histone H2B regulates nucleosome dynamics and access to DNA damage.
Rodriguez Y, Duan M, Wyrick JJ, Smerdon MJ. Rodriguez Y, et al. J Biol Chem. 2018 May 11;293(19):7376-7386. doi: 10.1074/jbc.RA117.000358. Epub 2018 Mar 27. J Biol Chem. 2018. PMID: 29588367 Free PMC article. - The structural location of DNA lesions in nucleosome core particles determines accessibility by base excision repair enzymes.
Rodriguez Y, Smerdon MJ. Rodriguez Y, et al. J Biol Chem. 2013 May 10;288(19):13863-75. doi: 10.1074/jbc.M112.441444. Epub 2013 Mar 29. J Biol Chem. 2013. PMID: 23543741 Free PMC article. - Challenges for base excision repair enzymes: Acquiring access to damaged DNA in chromatin.
Li C, Delaney S. Li C, et al. Enzymes. 2019;45:27-57. doi: 10.1016/bs.enz.2019.07.002. Epub 2019 Aug 9. Enzymes. 2019. PMID: 31627880 Review. - Base excision repair in nucleosome substrates.
Jagannathan I, Cole HA, Hayes JJ. Jagannathan I, et al. Chromosome Res. 2006;14(1):27-37. doi: 10.1007/s10577-005-1020-7. Epub 2006 Mar 3. Chromosome Res. 2006. PMID: 16506094 Review.
Cited by
- Structural basis for human OGG1 processing 8-oxodGuo within nucleosome core particles.
Ren M, Gut F, Fan Y, Ma J, Shan X, Yikilmazsoy A, Likhodeeva M, Hopfner KP, Zhou C. Ren M, et al. Nat Commun. 2024 Oct 31;15(1):9407. doi: 10.1038/s41467-024-53811-3. Nat Commun. 2024. PMID: 39477986 Free PMC article. - Eukaryotic Base Excision Repair: New Approaches Shine Light on Mechanism.
Beard WA, Horton JK, Prasad R, Wilson SH. Beard WA, et al. Annu Rev Biochem. 2019 Jun 20;88:137-162. doi: 10.1146/annurev-biochem-013118-111315. Annu Rev Biochem. 2019. PMID: 31220977 Free PMC article. Review. - Efficient cleavage of single and clustered AP site lesions within mono-nucleosome templates by CHO-K1 nuclear extract contrasts with retardation of incision by purified APE1.
Eccles LJ, Menoni H, Angelov D, Lomax ME, O'Neill P. Eccles LJ, et al. DNA Repair (Amst). 2015 Nov;35:27-36. doi: 10.1016/j.dnarep.2015.08.003. Epub 2015 Sep 12. DNA Repair (Amst). 2015. PMID: 26439176 Free PMC article. - The involvement of nucleotide excision repair proteins in the removal of oxidative DNA damage.
Kumar N, Raja S, Van Houten B. Kumar N, et al. Nucleic Acids Res. 2020 Nov 18;48(20):11227-11243. doi: 10.1093/nar/gkaa777. Nucleic Acids Res. 2020. PMID: 33010169 Free PMC article. Review. - VRK1 Depletion Facilitates the Synthetic Lethality of Temozolomide and Olaparib in Glioblastoma Cells.
Navarro-Carrasco E, Lazo PA. Navarro-Carrasco E, et al. Front Cell Dev Biol. 2021 Jun 14;9:683038. doi: 10.3389/fcell.2021.683038. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34195200 Free PMC article.
References
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. - PubMed
- Ausio J. Histone variants--the structure behind the function. Brief Funct Genomic Proteomic. 2006;5:228–243. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM008336/GM/NIGMS NIH HHS/United States
- R01 ES002614/ES/NIEHS NIH HHS/United States
- ES002614/ES/NIEHS NIH HHS/United States
- ES020955/ES/NIEHS NIH HHS/United States
- R01 ES004106/ES/NIEHS NIH HHS/United States
- ES004106/ES/NIEHS NIH HHS/United States
- R21 ES020955/ES/NIEHS NIH HHS/United States
- R37 ES002614/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources