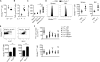IL-7-Induced Glycerol Transport and TAG Synthesis Promotes Memory CD8+ T Cell Longevity - PubMed (original) (raw)
IL-7-Induced Glycerol Transport and TAG Synthesis Promotes Memory CD8+ T Cell Longevity
Guoliang Cui et al. Cell. 2015.
Abstract
Memory T cells are critical for long-term immunity against reinfection and require interleukin-7 (IL-7), but the mechanisms by which IL-7 controls memory T cell survival, particularly metabolic fitness, remain elusive. We discover that IL-7 induces expression of the glycerol channel aquaporin 9 (AQP9) in virus-specific memory CD8+ T cells, but not naive cells, and that AQP9 is vitally required for their long-term survival. AQP9 deficiency impairs glycerol import into memory CD8+ T cells for fatty acid esterification and triglyceride (TAG) synthesis and storage. These defects can be rescued by ectopic expression of TAG synthases, which restores lipid stores and memory T cell survival. Finally, we find that TAG synthesis is a central component of IL-7-mediated survival of human and mouse memory CD8+T cells. This study uncovers the metabolic mechanisms by which IL-7 tailors the metabolism of memory T cells to promote their longevity and fast response to rechallenge.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
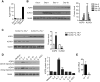
Figure 1. IL-7 Induces AQP9 Expression Selectively in Anti-viral Memory CD8+ T Cells and Their Precursors
(A–C) Naive, effector, and memory P14 CD8+ T cells were purified on the indicated dpiand theamount of Aqp9 mRNA was measured using DNA microarrays and analyzed by GeneSpring software (A) or protein using western blotting (B and C). (A) mRNA is normalized to naive samples. (B and C) Each lane represents an individual biological sample; Grp94 and KLRG1 were used as loading and internal monitoring controls, respectively. (C) KLRG1hiIL-7Rlo and KLRG1loIL-7Rhi effector CD8 T cell subsets were isolated at 14 dpi. The bar graphs on the right show densitometry quantification of the immunoblot bands. (D) P14 CD8+ T cells were primed with GP33–41 peptide for 3 days and then stimulated with various cytokines as indicated for 3 days before western blotting for AQP9. The bar graph on the right shows densitometry quantification of the immunoblot bands. (E) In vitro primed P14 CD8+ T cells were transferred to Il7+/+ or _Il7_−/− mice. At 7 days later, the donor cells were purified and Aqp9 mRNA levels were measured by qRT-PCR. Data in (B)–(E) are representative of two independent experiments (n = 3–6 mice/group): *p < 0.05 and **p < 0.01 (see also Figure S1).
Figure 2. AQP9 Deficiency Impairs Formation of LCMV-Specific Memory CD8+ T Cells
(A and B) Bone marrow chimeric mice containing a 1:1 ratio of Aqp9+/+ (open circles, Ly5.2+Thy1.1+) and _Aqp9_−/− (black squares, Ly5.2+Thy1.2+) bone marrow cells were infected with LCMV-Armstrong and the frequency of the two populations within the DbGP33–41-specific CD8+ T cells were analyzed longitudinally by flow cytometry. (C) _Aqp9_−/− or littermate Aqp9+/+ P14 CD8+ T cells (104 cells) were adoptively transferred into B6 mice that were subsequently infected with LCMV-Armstrong. The numbers of donor P14 CD8+ T cells were determined at the indicated dpi. (D) P14 chimeric mice described in (C) were given BrdU drinking water (1 mg/ml) from 30–51 dpi to measure the rates of homeostatic proliferation in the memory CD8+ T cells. Amounts of nuclear BrdU were measured by flow cytometry. (E) _Aqp9_−/− or littermate Aqp9+/+ P14 CD8+ T cells were purified at 25 dpi, labeled with CTV, adoptively transferred to naive B6 mice, and analyzed by flow cytometry for CTV dilution. The bar graph shows the percentages of divided cells. Data are representative (A, C–E) or cumulative (B) of three and four (C) independent experiments (n = 5–15 mice/group): **p < 0.01 (see also Figure S2).
Figure 3. AQP9 Deficiency Impairs TAG Synthesis and Storage in CD8+ T Cells
(A) _Aqp9_−/− or Aqp9+/+ P14 CD8+ T cells were cultured in vitro with GP33–41 peptide for 3 days and then in IL-7 for two days. The amount of free glycerol was measured in total cell lysates by a coupled enzymatic reaction system. (B) _Aqp9_−/− or Aqp9+/+ P14 memory CD8 T cells from 40 dpi were stained with the neutral lipid indicator Bodipy493/503 and analyzed by flow cytometry. (C) P14 CD8+ T cells described in (A) were pulsed with 0.1 μcurie per milliliter (Ci/ml) [1,2,3-14C]-Glycerol for 4 hr, then lipids were extracted and resolved by TLC. The bar graphs on the right show densitometry quantification of TAG and DAG autoradiography bands after a 2-week exposure. (D) P14 CD8+ T cells described in (A) were cultured in the presence or absence of glycerol (Gly) or OA for 2 days before lipid extraction and TLC assay. Standards for TAG and CHO were loaded on the left- and right-most lanes. The bar graph on the right shows densitometry quantification of TAG band. (E) Lipids were extracted from Aqp9+/+ and _Aqp9_−/− P14 CD8+ T cells described in (A) for LC-MS analysis of TAG isobaric species. Data are representative of two (B and E) and three (A, C, and D) independent experiments (n = 3–7 mice/group): *p < 0.05, **p < 0.01, and not significant (n.s.) (see also Figure S3).
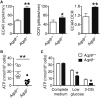
Figure 4. AQP9 Deficiency Reduces ATP Levels and Increases Glycolytic Rates in CD8+ T Cells
(A) _Aqp9_−/− or Aqp9+/+ P14 CD8+ T cells were cultured in vitro with GP33–41 peptide for 3 days and then in IL-7 for two days. Rates of ECAR and OCR were then measured using the Seahorse Extracellular Flux Analyzer. The bar graphs show the basal levels of ECAR, OCR, and the ratio between ECAR and OCR. (B) The amount of intracellular ATP was measured in _Aqp9_−/− or Aqp9+/+ P14 memory CD8 T cells from 40 dpi by a bioluminescence assay as described in Experimental Procedures. (C) P14 CD8+ T cells described in (A) were cultured in complete medium (11 millimolar [mM] glucose), low glucose medium (2.2 mM glucose), or complete medium plus the glycolysis inhibitor 2-DG and ATP levels were measured 12 hr later. Data are cumulative from three independent experiments (n = 8 mice/group) (B) or representative of three independent experiments (n = 3 mice/group) (A and C): *p < 0.05 and **p < 0.01.
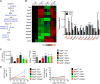
Figure 5. Overexpression of Glycerol Kinase and TAG Synthases Rescues Survival of _Aqp9_−/− Memory CD8+ T Cells
(A) Outline of TAG synthesis pathway with AQP9 and TAG synthases highlighted in blue. (B) Heat map shows the mean mRNA expression level of the indicated genes in virus-specific _Aqp9_−/− or Aqp9+/+ P14 CD8+ T cells purified at 0, 4.5, 8, and 40 dpi using qRT-PCR (values are normalized to naive CD8 T cells [day 0]). The bar graph shows the amount of mRNA relative to L9 mRNA in memory CD8 T cells (40 dpi) and the statistically significant differentially expressed genes are highlighted in red. (C) _Aqp9_−/− P14 CD8+ T cells were transduced with RVs overexpressing _Gyk, Dgat1, Gpat_1, Mogat1, or control EV and adoptively transferred into B6 mice that were subsequently infected with LCMV-Armstrong. Aqp9+/+ P14 CD8 T cells transduced with EV were included for comparison (dark gray). The bar graphs show the numbers of donor P14 CD8+ T cells and MFI of Bodipy493/503 staining at 40 dpi. (D) Longitudinal analysis of the RV-transduced _Aqp9_−/− and Aqp9+/+ P14 CD8+ T cells, as described in (C). At each time point, the frequency of RV+ cells was normalized to EV control cells and plotted in the line graph. Data are cumulative of three (B–D) experiments (n = 6–10 mice/group): *p < 0.05, **p < 0.01, and not significant (n.s.).
Figure 6. IL-7-Driven Glycerol Metabolism and TAG Synthesis Are Critical for Memory CD8+ T Cell Survival
(A and B) Purified memory P14 CD8+ T cells from 50–70 dpi were (A) transferred to Il7+/+ and _Il7_−/− mice and 5 days later the donor cells were stained with Bodipy493/503 and analyzed by flow cytometry or (B) stimulated with IL-7 for 12 hr and then TAG levels were measured as described in Experimental Procedures. (C) Purified naive or memory P14 CD8+ T cells (from 50–70 dpi) were stimulated with IL-7 for 12 hr and then TAG levels were measured using Bodipy493/503 staining and flow cytometry. (D) P14 chimeric mice containing memory P14 CD8+ T cells from 40–60 dpi were injected with IL-7/anti IL-7 (M25) complex. At 3 days later, TAG levels in donor P14 cells in the spleen and bone marrow were measured using Bodipy493/503 staining and flow cytometry. (E) Memory P14 CD8+ T cells purified at 50–150 dpi were stained with Bodipy493/503, Ki-67, and Bcl-2 and analyzed by flow cytometry. The bar graphs show Bodipy493/503 MFI in the indicated cell populations. (F) Memory P14 CD8+ T cells (from 30–40 dpi) overexpressing Gyk, Dgat1, Gpat1, Mogat1, or EV were adoptively transferred into _Il7_−/− mice. For comparison, the EV-transduced cells were also transferred into Il7+/+ mice (open circles). At 2–4 weeks later, donor RV+ P14 CD8+ T cells were enumerated and stained with Bodipy493/503. Values were normalized to those of EV-transduced cells in Il7+/+ mice (open circles). Data shown are cumulative of two (D), three (A–C), and four (F) independent experiments or representative of five independent experiments (E) (n = 5–20 mice/group): *p < 0.05 and **p < 0.01 (see also Figure S4).
Figure 7. IL-7 Drives TAG Synthesis and Promotes Human Memory CD8+ T Cell Survival
(A and B) CCR7+ CD45RA+ CD8+ naive T cells, CCR7− CD45RA+ CD8+ TEMRA, CCR7− CD45RA− CD8+ TEM cells, and CCR7+ CD45RA− CD8+ TCM cells were FACS purified and Aqp9 mRNA expression was measured by qRT-PCR (values shown are normalized to naive T cells). (C) Scatter plot shows Aqp9 mRNA expression in CD45RA− CD8+ T cells stimulated with IL-7 for 24 hr, measured by qRT-PCR, and normalized to the untreated control cells. (D) Freshly isolated human CD45RA− CD8+ T cells were stimulated with or without IL-7 or IL-7 plus glycerol and OA for 24 hr before Bodipy493/503 staining. Bodipy493/503 MFI was normalized to the untreated control cells. (E) Freshly isolated human CD45RA− CD8+ T cells from two individuals were pulsed with 0.1 μCi/ml [1,2,3-14C]-Glycerol in the absence or presence of IL-7 or the indicated drugs for 4 hr before lipid extraction and TLC assay. The bar graphs on the right show densitometry quantification of TAG and PL autoradiography bands after a 10-week exposure. (F) Histograms show the percentage of Annexin V+ CD45RA− CD8+ T cells after a 12 hr treatment as indicated. The bar graph on the right shows the cumulative data of six samples. Data shown are a cumulative of three (B–D) independent experiments or representative of three (E and F) independent experiments (n = 5–8 subjects/group): *p < 0.05 and **p < 0.01.
Similar articles
- Immunology. CD8alphaalpha and T cell memory.
Kim SV, Flavell RA. Kim SV, et al. Science. 2004 Apr 23;304(5670):529-30. doi: 10.1126/science.1097678. Science. 2004. PMID: 15105485 No abstract available. - Distinct roles for IL-2 and IL-15 in the differentiation and survival of CD8+ effector and memory T cells.
Mitchell DM, Ravkov EV, Williams MA. Mitchell DM, et al. J Immunol. 2010 Jun 15;184(12):6719-30. doi: 10.4049/jimmunol.0904089. Epub 2010 May 14. J Immunol. 2010. PMID: 20483725 Free PMC article. - Signal 3 requirement for memory CD8+ T-cell activation is determined by the infectious pathogen.
Keppler SJ, Aichele P. Keppler SJ, et al. Eur J Immunol. 2011 Nov;41(11):3176-86. doi: 10.1002/eji.201141537. Epub 2011 Sep 26. Eur J Immunol. 2011. PMID: 21830209 - Decisions on the road to memory.
Amsen D, Backer RA, Helbig C. Amsen D, et al. Adv Exp Med Biol. 2013;785:107-20. doi: 10.1007/978-1-4614-6217-0_12. Adv Exp Med Biol. 2013. PMID: 23456843 Review. - Fatty acid metabolism in CD8+ T cell memory: Challenging current concepts.
Raud B, McGuire PJ, Jones RG, Sparwasser T, Berod L. Raud B, et al. Immunol Rev. 2018 May;283(1):213-231. doi: 10.1111/imr.12655. Immunol Rev. 2018. PMID: 29664569 Free PMC article. Review.
Cited by
- Tissue-Specific Contributions to Control of T Cell Immunity.
Poholek AC. Poholek AC. Immunohorizons. 2021 Jun 8;5(6):410-423. doi: 10.4049/immunohorizons.2000103. Immunohorizons. 2021. PMID: 34103371 Free PMC article. - Aquaporins in Immune Cells and Inflammation: New Targets for Drug Development.
da Silva IV, Soveral G. da Silva IV, et al. Int J Mol Sci. 2021 Feb 12;22(4):1845. doi: 10.3390/ijms22041845. Int J Mol Sci. 2021. PMID: 33673336 Free PMC article. Review. - Immunometabolism of regulatory T cells.
Newton R, Priyadharshini B, Turka LA. Newton R, et al. Nat Immunol. 2016 May 19;17(6):618-25. doi: 10.1038/ni.3466. Nat Immunol. 2016. PMID: 27196520 Free PMC article. Review. - Comparison of IL-2 vs IL-7/IL-15 for the generation of NY-ESO-1-specific T cells.
Gong W, Hoffmann JM, Stock S, Wang L, Liu Y, Schubert ML, Neuber B, Hückelhoven-Krauss A, Gern U, Schmitt A, Müller-Tidow C, Shiku H, Schmitt M, Sellner L. Gong W, et al. Cancer Immunol Immunother. 2019 Jul;68(7):1195-1209. doi: 10.1007/s00262-019-02354-4. Epub 2019 Jun 8. Cancer Immunol Immunother. 2019. PMID: 31177329 Free PMC article. - Directing T-Cell Immune Responses for Cancer Vaccination and Immunotherapy.
Smith PL, Piadel K, Dalgleish AG. Smith PL, et al. Vaccines (Basel). 2021 Nov 25;9(12):1392. doi: 10.3390/vaccines9121392. Vaccines (Basel). 2021. PMID: 34960140 Free PMC article. Review.
References
- Abrami L, Berthonaud V, Deen PM, Rousselet G, Tacnet F, Ripoche P. Glycerol permeability of mutant aquaporin 1 and other AQP-MIP proteins: inhibition studies. Pflugers Arch. 1996;431:408–414. - PubMed
- Akashi K, Kondo M, von Freeden-Jeffry U, Murray R, Weissman IL. Bcl-2 rescues T lymphopoiesis in interleukin-7 receptor-deficient mice. Cell. 1997;89:1033–1041. - PubMed
- Barra NG, Reid S, MacKenzie R, Werstuck G, Trigatti BL, Richards C, Holloway AC, Ashkar AA. Interleukin-15 contributes to the regulation of murine adipose tissue and human adipocytes. Obesity (Silver Spring) 2010;18:1601–1607. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32GM07205/GM/NIGMS NIH HHS/United States
- R01 AI074699/AI/NIAID NIH HHS/United States
- U19AI089992/AI/NIAID NIH HHS/United States
- R37AI066232/R01AI074699/AI/NIAID NIH HHS/United States
- R37 AI066232/AI/NIAID NIH HHS/United States
- T32 GM007205/GM/NIGMS NIH HHS/United States
- Howard Hughes Medical Institute/United States
- U19 AI089992/AI/NIAID NIH HHS/United States
- R01 AI066232/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials