A microfluidic device for label-free, physical capture of circulating tumor cell clusters - PubMed (original) (raw)
doi: 10.1038/nmeth.3404. Epub 2015 May 18.
Nicola Aceto 2, Nikola Kojic 3, Maria C Donaldson 4, Mahnaz Zeinali 5, Bashar Hamza 6, Amanda Engstrom 4, Huili Zhu 4, Tilak K Sundaresan 4, David T Miyamoto 7, Xi Luo 4, Aditya Bardia 2, Ben S Wittner 4, Sridhar Ramaswamy 2, Toshi Shioda 4, David T Ting 2, Shannon L Stott 8, Ravi Kapur 6, Shyamala Maheswaran 9, Daniel A Haber 10, Mehmet Toner 3
Affiliations
- PMID: 25984697
- PMCID: PMC4490017
- DOI: 10.1038/nmeth.3404
A microfluidic device for label-free, physical capture of circulating tumor cell clusters
A Fatih Sarioglu et al. Nat Methods. 2015 Jul.
Abstract
Cancer cells metastasize through the bloodstream either as single migratory circulating tumor cells (CTCs) or as multicellular groupings (CTC clusters). Existing technologies for CTC enrichment are designed to isolate single CTCs, and although CTC clusters are detectable in some cases, their true prevalence and significance remain to be determined. Here we developed a microchip technology (the Cluster-Chip) to capture CTC clusters independently of tumor-specific markers from unprocessed blood. CTC clusters are isolated through specialized bifurcating traps under low-shear stress conditions that preserve their integrity, and even two-cell clusters are captured efficiently. Using the Cluster-Chip, we identified CTC clusters in 30-40% of patients with metastatic breast or prostate cancer or with melanoma. RNA sequencing of CTC clusters confirmed their tumor origin and identified tissue-derived macrophages within the clusters. Efficient capture of CTC clusters will enable the detailed characterization of their biological properties and role in metastasis.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests. A.F.S and M.T. are inventors on a patent MGH filed to protect the Cluster-Chip technology.
Figures
Figure 1
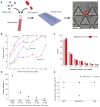
The design and operation of the Cluster-Chip (a) Schematic representation of the Cluster-Chip operation. Cluster-Chip captures CTC clusters from unprocessed whole blood while single cells pass through. (b) SEM micrographs of the Cluster-Chip showing multiple rows of shifted triangular pillars forming consecutive cluster traps (left) and a high magnification image of a cluster trap (right). Scale bars 60 μm. (c) A full image of the Cluster-Chip. The blood from a single inlet is uniformly distributed over 4096 parallel trapping paths and then collected to a single outlet. The inset shows a close up of a CTC cluster-trapping region with part of the microfluidic distribution and collection networks. (d) A spiked 2-cell LNCaP cluster captured on the Cluster-Chip (top) and schematic explaining the dynamic balance that holds it captured (bottom). Forces acting on the cell cluster are drag forces (_F_D) due to fluid flow, reaction forces (_F_R) from the pillars and frictional forces (_F_F) including the effect of cell adhesion. (e) Finite element analysis comparing the cell cluster dynamics in the Cluster-Chip (left) and in a filter with half the opening size (right). The diameters of individual cells are 15 μm and the opening width (w) is 12 μm.
Figure 2
Characterization of the Cluster-Chip capture using cell lines spiked in whole blood (a) The schematic diagram of the experimental procedure used to ensure against artificial cell aggregation. (b) Cluster-Chip cell cluster capture rate measured using artificial clusters of MDA-MB-231 cell line spiked in whole blood. Capture rate is shown as a function of the number of cells in the cluster at different flow rates. (c) Histograms of the number of cells in the cell clusters within spiked population and cell clusters captured in the Cluster-Chip. Error bars show s.e.m from three independent experiments. (d) Comparison of cluster capture efficiency of Cluster-Chip with membrane filters operated under different pressures using human breast cancer cell line MDA-MB-231. Effective whole blood processing rate for each condition is noted in parentheses on the x-axis. Error bars show s.e.m from three independent experiments. (e) Comparison of cluster capture efficiency of Cluster-Chip with immunoaffinity-based HB-Chip using three human breast cancer cell lines. Surface EpCAM expression is highest in MCF7 and lowest in LBX1. Cluster-Chip has higher cluster capture efficiency for all cases. Error bars show s.e.m from three independent experiments. Scale bars 60 μm.
Figure 3
Release of captured clusters from the Cluster-Chip (a) Schematic diagram of the experimental setup (top) and individual steps of the CTC cluster release process (bottom). The bulk of the blood sample is continuously rocked at room temperature and is cooled only when it is being processed by the Cluster-Chip. (b) Release efficiency of MDA-MB-231 clusters from the chip as a function of the reverse flow rate and the processing temperature. (c) Nonspecific binding of leukocytes on the Cluster-Chip when the sample is processed at room temperature (left) and at 4°C (right). The leukocytes were fixed with 4% PFA and stained with DAPI. Fluorescent images were overlaid on bright field images. (d) Images of the product released in solution from the Cluster-Chip operated at room temperature (top-left) and at 4°C (bottom-left). Relative purity of released cell clusters against contaminating blood cells when Cluster-Chip is operated at room temperature and 4°C (right). Scale bars 60 μm.
Figure 4
Capture of CTC-clusters from blood samples of patients with metastatic cancer (a) Representative images of CTC-clusters isolated form patients with metastatic breast cancer: (Left) Brightfield and fluorescent images of a live CTC-cluster stained for common breast cancer surface markers; (Middle) SEM micrograph of a fixed CTC-cluster; (Right) Fluorescent image of a highly deformable CTC-cluster stained for cytokeratin. Note that this CTC-cluster is not split but is highly strained even under slow flow in the Cluster-Chip. This particular case clearly justifies the need for the elasticity-independent capture mechanism of the Cluster-Chip. (b) Percentage of patients with CTC-clusters in breast, melanoma and prostate cancer. (c) Size distribution of CTC-clusters isolated from breast, melanoma and prostate cancer patients. The box plots show the 25th, 50th and 75th percentile for each disease type. Scale bars 20 μm.
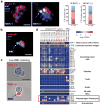
Figure 5
Immunocytochemical and molecular characterization of patient CTC-clusters (a) Images of a Ki67-negative and a Ki67-positive CTC-clusters stained with CK (red), Ki67 (yellow), CD45 (green) and DAPI (nuclei, blue). The bar graphs show the percentage of Ki67-positive CTC-clusters in this patient (left; n=21) and the percentage of Ki67-positive cells within CTC-clusters (right; n=64). (b) Image of a CTC-cluster associated to a white blood cell (WBC). The cells were stained with CK (red), CD45 (green) and DAPI (nuclei, blue). (c) Images of WBC-negative (top) and WBC-positive (bottom) CTC-clusters released from the Cluster-Chip and live-stained with TexasRed-conjugated antibodies against CD45, CD14 and CD16 (red). (d) Heatmap showing expression levels of transcripts associated to CTCs, macrophages/monocytes, T cells, B cells, natural killer (NK) cells, hematopoietic stem cells (HSCs), granulocytes and platelets in 15 CTC-clusters isolated at a single timepoint from a patient with metastatic breast cancer. RPM: reads per million. Scale bars 20 μm
Similar articles
- Diagnostic microchip to assay 3D colony-growth potential of captured circulating tumor cells.
Bichsel CA, Gobaa S, Kobel S, Secondini C, Thalmann GN, Cecchini MG, Lutolf MP. Bichsel CA, et al. Lab Chip. 2012 Jul 7;12(13):2313-6. doi: 10.1039/c2lc40130d. Epub 2012 May 8. Lab Chip. 2012. PMID: 22565166 - Prospective assessment of the prognostic value of circulating tumor cells and their clusters in patients with advanced-stage breast cancer.
Mu Z, Wang C, Ye Z, Austin L, Civan J, Hyslop T, Palazzo JP, Jaslow R, Li B, Myers RE, Jiang J, Xing J, Yang H, Cristofanilli M. Mu Z, et al. Breast Cancer Res Treat. 2015 Dec;154(3):563-71. doi: 10.1007/s10549-015-3636-4. Epub 2015 Nov 16. Breast Cancer Res Treat. 2015. PMID: 26573830 - Liquid biopsy using the nanotube-CTC-chip: capture of invasive CTCs with high purity using preferential adherence in breast cancer patients.
Loeian MS, Mehdi Aghaei S, Farhadi F, Rai V, Yang HW, Johnson MD, Aqil F, Mandadi M, Rai SN, Panchapakesan B. Loeian MS, et al. Lab Chip. 2019 Jun 7;19(11):1899-1915. doi: 10.1039/c9lc00274j. Epub 2019 May 3. Lab Chip. 2019. PMID: 31049504 - Relevance of CTC Clusters in Breast Cancer Metastasis.
Piñeiro R, Martínez-Pena I, López-López R. Piñeiro R, et al. Adv Exp Med Biol. 2020;1220:93-115. doi: 10.1007/978-3-030-35805-1_7. Adv Exp Med Biol. 2020. PMID: 32304082 Review. - Circulating tumor cell clusters: What we know and what we expect (Review).
Hong Y, Fang F, Zhang Q. Hong Y, et al. Int J Oncol. 2016 Dec;49(6):2206-2216. doi: 10.3892/ijo.2016.3747. Epub 2016 Oct 24. Int J Oncol. 2016. PMID: 27779656 Free PMC article. Review.
Cited by
- Trapping of a Single Microparticle Using AC Dielectrophoresis Forces in a Microfluidic Chip.
Wang Y, Tong N, Li F, Zhao K, Wang D, Niu Y, Xu F, Cheng J, Wang J. Wang Y, et al. Micromachines (Basel). 2023 Jan 8;14(1):159. doi: 10.3390/mi14010159. Micromachines (Basel). 2023. PMID: 36677221 Free PMC article. - [Recent advances in isolation and detection of circulating tumor cells with a microfluidic system].
Cao R, Zhang M, Yu H, Qin J. Cao R, et al. Se Pu. 2022 Mar 8;40(3):213-223. doi: 10.3724/SP.J.1123.2021.07009. Se Pu. 2022. PMID: 35243831 Free PMC article. Review. Chinese. - Ex vivo expansion of circulating tumour cells (CTCs).
Mohamed BM, Ward MP, Bates M, Spillane CD, Kelly T, Martin C, Gallagher M, Heffernan S, Norris L, Kennedy J, Saadeh FA, Gleeson N, Brooks DA, Brooks RD, Selemidis S, O'Toole S, O'Leary JJ. Mohamed BM, et al. Sci Rep. 2023 Mar 6;13(1):3704. doi: 10.1038/s41598-023-30733-6. Sci Rep. 2023. PMID: 36879003 Free PMC article. - The Continuous Concentration of Particles and Cancer Cell Line Using Cell Margination in a Groove-Based Channel.
Yan S, Yuan D, Zhao Q, Zhang J, Li W. Yan S, et al. Micromachines (Basel). 2017 Oct 25;8(11):315. doi: 10.3390/mi8110315. Micromachines (Basel). 2017. PMID: 30400505 Free PMC article. - Optofluidic real-time cell sorter for longitudinal CTC studies in mouse models of cancer.
Hamza B, Ng SR, Prakadan SM, Delgado FF, Chin CR, King EM, Yang LF, Davidson SM, DeGouveia KL, Cermak N, Navia AW, Winter PS, Drake RS, Tammela T, Li CM, Papagiannakopoulos T, Gupta AJ, Shaw Bagnall J, Knudsen SM, Vander Heiden MG, Wasserman SC, Jacks T, Shalek AK, Manalis SR. Hamza B, et al. Proc Natl Acad Sci U S A. 2019 Feb 5;116(6):2232-2236. doi: 10.1073/pnas.1814102116. Epub 2019 Jan 23. Proc Natl Acad Sci U S A. 2019. PMID: 30674677 Free PMC article.
References
- Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nature Reviews Cancer. 2008;8:329–340. - PubMed
Publication types
MeSH terms
Grants and funding
- U01 EB012493/EB/NIBIB NIH HHS/United States
- R01 EB008047/EB/NIBIB NIH HHS/United States
- Howard Hughes Medical Institute/United States
- K12 CA087723/CA/NCI NIH HHS/United States
- P41 EB002503/EB/NIBIB NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases